We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:R. Jeremy Johnson/Mitochondrial Calcium Uniporter
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
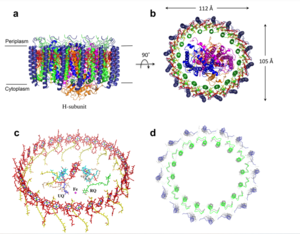
[[Image:structure.png|300 px|right|thumb|Figure 1: Structure of mitochondrial calcium uniporter colored by functional domain. The transmembrane domain is highlighted in salmon, the matrix domain in light cyan, coiled-coil domain in dark violet, and the N-terminal domain in slate blue. [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] | [[Image:structure.png|300 px|right|thumb|Figure 1: Structure of mitochondrial calcium uniporter colored by functional domain. The transmembrane domain is highlighted in salmon, the matrix domain in light cyan, coiled-coil domain in dark violet, and the N-terminal domain in slate blue. [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] | ||
===Mitochondrial Calcium Uniporter Complex=== | ===Mitochondrial Calcium Uniporter Complex=== | ||
| - | |||
The mitochondrial calcium uniporter complex exists as a large complex (around 480 kDa in humans) made up of both pore-forming and regulatory subunits.<ref name="Wang"/> The MCU is a complex composed of regulatory subunits including mitochondrial calcium uptake (MICU), essential MICU regulator (EMRE), MCU regulatory subunit b (MCUb), and MCU regulator 1 (MCUR1). <ref name="Fan" /> The mitochondrial uptake proteins (MICU1 and MICU2) are regulatory proteins in the MCU complex that exist in the IMS and contain [https://en.wikipedia.org/wiki/EF_hand EF hand domains] for calcium binding to control transport through the channel of the MCU complex.<ref name="Wang"/> When calcium ion concentration in the IMS is low, MICU1 and 2 block the MCU to prevent uptake of calcium.<ref name="Wang"/> In the presence of high calcium concentrations, more calcium binds to these regulatory proteins and they undergo a conformational change to allow calcium ions through the MCU and into the matrix.<ref name="Wang"/> In fact, when calcium levels are below 500 nM, MICU1 can block movement of calcium by itself, calcium levels between 500 nM and 1,500 nM require both MICU1 and MICU2 to block ion entry, and any concentration over 1,500 nM is sufficient for calcium entry.<ref name="Giorgi"/> Another regulatory protein, MCUR1 is a cofactor in the assembly of the [https://en.wikipedia.org/wiki/Electron_transport_chain respiratory chain] rather than an essential part of the uniporter.<ref name="Giorgi"/> Though the MCU is able to take up calcium independently, there are two other pore-forming subunits, the MCUb and the essential MCU regulator (EMRE).<ref name="Wang"/> MCUb is similar to MCU, but certain amino acids differ and make it an inhibitory subunit.<ref name="Wang"/> The EMRE is located in the IMS and connects MICU1 and MICU2 to the MCU.<ref name="Giorgi"/> It also contributes to regulation of calcium intake in the MCU.<ref name="Wang"/> | The mitochondrial calcium uniporter complex exists as a large complex (around 480 kDa in humans) made up of both pore-forming and regulatory subunits.<ref name="Wang"/> The MCU is a complex composed of regulatory subunits including mitochondrial calcium uptake (MICU), essential MICU regulator (EMRE), MCU regulatory subunit b (MCUb), and MCU regulator 1 (MCUR1). <ref name="Fan" /> The mitochondrial uptake proteins (MICU1 and MICU2) are regulatory proteins in the MCU complex that exist in the IMS and contain [https://en.wikipedia.org/wiki/EF_hand EF hand domains] for calcium binding to control transport through the channel of the MCU complex.<ref name="Wang"/> When calcium ion concentration in the IMS is low, MICU1 and 2 block the MCU to prevent uptake of calcium.<ref name="Wang"/> In the presence of high calcium concentrations, more calcium binds to these regulatory proteins and they undergo a conformational change to allow calcium ions through the MCU and into the matrix.<ref name="Wang"/> In fact, when calcium levels are below 500 nM, MICU1 can block movement of calcium by itself, calcium levels between 500 nM and 1,500 nM require both MICU1 and MICU2 to block ion entry, and any concentration over 1,500 nM is sufficient for calcium entry.<ref name="Giorgi"/> Another regulatory protein, MCUR1 is a cofactor in the assembly of the [https://en.wikipedia.org/wiki/Electron_transport_chain respiratory chain] rather than an essential part of the uniporter.<ref name="Giorgi"/> Though the MCU is able to take up calcium independently, there are two other pore-forming subunits, the MCUb and the essential MCU regulator (EMRE).<ref name="Wang"/> MCUb is similar to MCU, but certain amino acids differ and make it an inhibitory subunit.<ref name="Wang"/> The EMRE is located in the IMS and connects MICU1 and MICU2 to the MCU.<ref name="Giorgi"/> It also contributes to regulation of calcium intake in the MCU.<ref name="Wang"/> | ||
===Mitochondrial Calcium Uniporter=== | ===Mitochondrial Calcium Uniporter=== | ||
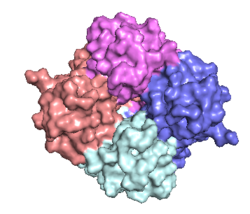
The <scene name='83/832952/Starting_scene/5'>mitochondrial calcium uniporter (MCU)</scene> is the ion channel component (Figure 1). The MCU was originally thought to be composed of [https://en.wikipedia.org/wiki/Pentamer pentamer] of five identical subunits, but it is now known to exist as a [https://en.wikipedia.org/wiki/Dimer_(chemistry) dimer] of <scene name='83/832952/Dimer_of_dimers/5'>dimers</scene> (Figure 2).<ref name="Woods">PMID:31869674</ref> The <scene name='83/837230/Ntermsymmetry/1'>dimeric</scene> organization of the MCU is described as a <scene name='83/837230/Pyramid/2'>tetrameric truncated pyramid</scene>. The protein is composed of a <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene>, a <scene name='83/837230/Coiled_coil/3'>coiled coil domain</scene>, and a <scene name='83/837230/Nterm/2'>N-Terminal Domain</scene> (NTD) (Figure 1).<ref name="Woods"/> The hydrophobic <scene name='83/832952/New_ones/3'>transmembrane domain</scene> is located in the inner mitochondrial membrane ([https://en.wikipedia.org/wiki/Inner_mitochondrial_membrane IMM]) and the hydrophilic coiled-coil domain exists in the mitochondrial matrix.<ref name="Baradaran"/> | The <scene name='83/832952/Starting_scene/5'>mitochondrial calcium uniporter (MCU)</scene> is the ion channel component (Figure 1). The MCU was originally thought to be composed of [https://en.wikipedia.org/wiki/Pentamer pentamer] of five identical subunits, but it is now known to exist as a [https://en.wikipedia.org/wiki/Dimer_(chemistry) dimer] of <scene name='83/832952/Dimer_of_dimers/5'>dimers</scene> (Figure 2).<ref name="Woods">PMID:31869674</ref> The <scene name='83/837230/Ntermsymmetry/1'>dimeric</scene> organization of the MCU is described as a <scene name='83/837230/Pyramid/2'>tetrameric truncated pyramid</scene>. The protein is composed of a <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene>, a <scene name='83/837230/Coiled_coil/3'>coiled coil domain</scene>, and a <scene name='83/837230/Nterm/2'>N-Terminal Domain</scene> (NTD) (Figure 1).<ref name="Woods"/> The hydrophobic <scene name='83/832952/New_ones/3'>transmembrane domain</scene> is located in the inner mitochondrial membrane ([https://en.wikipedia.org/wiki/Inner_mitochondrial_membrane IMM]) and the hydrophilic coiled-coil domain exists in the mitochondrial matrix.<ref name="Baradaran"/> | ||
| - | [[Image:Nterm.png| | + | [[Image:Nterm.png|250 px|right|thumb|Figure 2: Symmetry and organization of subunits from looking down into the uniporter from the inner mitochondrial membrane [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] |
===Transmembrane Domain=== | ===Transmembrane Domain=== | ||
The <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene> is on the [https://en.wikipedia.org/wiki/Mitochondrion#Structure inner mitochondrial membrane] open to the inner membrane space (Figure 1). The <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene> consists of eight separate helices (TM1 and TM2 from each subunit) that are connected by mostly hydrophobic amino acids in the IMS and has <scene name='83/832952/Starting_scene/5'>four-fold</scene> symmetry (Figure 4).<ref name="Baradaran"/> The small <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> pore, highly specific for calcium binding is located in <scene name='83/837230/Tm2/1'>transmembrane 2</scene> (TM2) while <scene name='83/837230/Transmembrane_1/2'>transmembrane 1</scene> (TM1) surrounds the pore. <scene name='83/832952/Tm1/2'>TM1</scene> packs tightly against <scene name='83/832952/Tm2/2'>TM2</scene> from the neighboring subunit which conveys a sense of domain-swapping.<ref name="Fan">PMID:29995856</ref> The transmembrane domain of MCU can be roughly divided into a narrow outer leaflet portion with a <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene>, which is lined by the <scene name='83/832952/Tm2/2'>TM2</scene> helices and a wide inner leaflet.<ref name="Baradaran"/> | The <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene> is on the [https://en.wikipedia.org/wiki/Mitochondrion#Structure inner mitochondrial membrane] open to the inner membrane space (Figure 1). The <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene> consists of eight separate helices (TM1 and TM2 from each subunit) that are connected by mostly hydrophobic amino acids in the IMS and has <scene name='83/832952/Starting_scene/5'>four-fold</scene> symmetry (Figure 4).<ref name="Baradaran"/> The small <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> pore, highly specific for calcium binding is located in <scene name='83/837230/Tm2/1'>transmembrane 2</scene> (TM2) while <scene name='83/837230/Transmembrane_1/2'>transmembrane 1</scene> (TM1) surrounds the pore. <scene name='83/832952/Tm1/2'>TM1</scene> packs tightly against <scene name='83/832952/Tm2/2'>TM2</scene> from the neighboring subunit which conveys a sense of domain-swapping.<ref name="Fan">PMID:29995856</ref> The transmembrane domain of MCU can be roughly divided into a narrow outer leaflet portion with a <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene>, which is lined by the <scene name='83/832952/Tm2/2'>TM2</scene> helices and a wide inner leaflet.<ref name="Baradaran"/> | ||
| Line 28: | Line 27: | ||
===Selectivity Filter=== | ===Selectivity Filter=== | ||
| - | [[Image:Electronegativity_MCU_4.jpg| | + | [[Image:Electronegativity_MCU_4.jpg|250 px|right|thumb|Figure3: Electronegativity of the MCU viewed from outside the channel. The high concentration of negative charge (shown in red) attracts the positive calcium ions. [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] |
The <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> of the MCU is composed by many acidic amino acids near the narrow mouth of the channel which leads to high affinity for calcium ([https://en.wikipedia.org/wiki/Dissociation_constant dissociation constant] of less than 2nM) (Figure 3).<ref name="Baradaran"/> Negatively charged aspartates <scene name='83/832952/New_ones/2'>(Asp221)</scene> at the mouth of the MCU congregate positively charged <scene name='83/832952/Calcium/4'>calcium ions</scene> at the entrance of the channel.<ref name="Baradaran"/> A highly conserved <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> [https://en.wikipedia.org/wiki/Sequence_motif motif] in the TM2 helices form the selectivity pore which selects for calcium transport over other similar ions.<ref name="Baradaran"/> | The <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> of the MCU is composed by many acidic amino acids near the narrow mouth of the channel which leads to high affinity for calcium ([https://en.wikipedia.org/wiki/Dissociation_constant dissociation constant] of less than 2nM) (Figure 3).<ref name="Baradaran"/> Negatively charged aspartates <scene name='83/832952/New_ones/2'>(Asp221)</scene> at the mouth of the MCU congregate positively charged <scene name='83/832952/Calcium/4'>calcium ions</scene> at the entrance of the channel.<ref name="Baradaran"/> A highly conserved <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> [https://en.wikipedia.org/wiki/Sequence_motif motif] in the TM2 helices form the selectivity pore which selects for calcium transport over other similar ions.<ref name="Baradaran"/> | ||
Revision as of 19:44, 8 May 2020
Mitochondrial Calcium Uniporter (MCU) Complex
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 1.22 1.23 1.24 1.25 1.26 1.27 Baradaran R, Wang C, Siliciano AF, Long SB. Cryo-EM structures of fungal and metazoan mitochondrial calcium uniporters. Nature. 2018 Jul 11. pii: 10.1038/s41586-018-0331-8. doi:, 10.1038/s41586-018-0331-8. PMID:29995857 doi:http://dx.doi.org/10.1038/s41586-018-0331-8
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 2.18 Woods JJ, Wilson JJ. Inhibitors of the mitochondrial calcium uniporter for the treatment of disease. Curr Opin Chem Biol. 2019 Dec 20;55:9-18. doi: 10.1016/j.cbpa.2019.11.006. PMID:31869674 doi:http://dx.doi.org/10.1016/j.cbpa.2019.11.006
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Giorgi C, Marchi S, Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol. 2018 Nov;19(11):713-730. doi: 10.1038/s41580-018-0052-8. PMID:30143745 doi:http://dx.doi.org/10.1038/s41580-018-0052-8
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 4.13 4.14 4.15 4.16 4.17 Wang CH, Wei YH. Role of mitochondrial dysfunction and dysregulation of Ca(2+) homeostasis in the pathophysiology of insulin resistance and type 2 diabetes. J Biomed Sci. 2017 Sep 7;24(1):70. doi: 10.1186/s12929-017-0375-3. PMID:28882140 doi:http://dx.doi.org/10.1186/s12929-017-0375-3
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 Fan C, Fan M, Orlando BJ, Fastman NM, Zhang J, Xu Y, Chambers MG, Xu X, Perry K, Liao M, Feng L. X-ray and cryo-EM structures of the mitochondrial calcium uniporter. Nature. 2018 Jul 11. pii: 10.1038/s41586-018-0330-9. doi:, 10.1038/s41586-018-0330-9. PMID:29995856 doi:http://dx.doi.org/10.1038/s41586-018-0330-9
- ↑ Yoo J, Wu M, Yin Y, Herzik MA Jr, Lander GC, Lee SY. Cryo-EM structure of a mitochondrial calcium uniporter. Science. 2018 Jun 28. pii: science.aar4056. doi: 10.1126/science.aar4056. PMID:29954988 doi:http://dx.doi.org/10.1126/science.aar4056
Student Contributors
Ryan Heumann
Lizzy Ratz
Holly Rowe
Madi Summers
Rieser Wells