User:Lukáš Cakl/Sandbox 1
From Proteopedia
(Difference between revisions)
m |
m |
||
| Line 16: | Line 16: | ||
[[Image:Tyr133_difference.png|thumb|hPCNA-S228I disorder tyr133 difference]] | [[Image:Tyr133_difference.png|thumb|hPCNA-S228I disorder tyr133 difference]] | ||
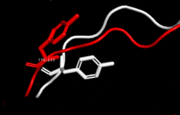
A disease causing variant in human, which causes a large conformational change of the PIP binding pocket. While the overal structure of the S228I mutant is mostly similar to the wild type, the change causes displacements extending from the mutation site, which affect the IDCL. Notably Tyr133, a highly conservated residue, rotates outwards by nearly 90° to prevent spherical conflicts with Ile228. The changes cause the binding cavity of S228I to be about a third of the size of its wild type equivalent and makes it incompetent for PIP binding. Such changes, if they were permanent, would however be lethal, as the removal of PCNA-FEN1 interactions is lethal on its own <ref>DOI:10.1158/0008-5472.CAN-08-0168</ref>. While some client proteins (e.q. [[p21<sup>CIP1</sup>]]) are left relatively unaffected even in this state, others' binding energetics are hugely disrupted (e.q. [[RNase H2B]] or [[FEN1]]). The explaination for survival of an individual carrying this mutation is sufficient malleability of IDCL (and thus of PCNA itself). Thus a PIP might be able to initiate an "induced-fit". Even here the mutant is disadvantaged as the wild type has B-factors ~40% higher than the mutant. Thus the mutation negatively affects even the IDCL dynamics. <ref>PMID:26688547</ref>. <scene name='84/842997/Fen1_with_pcna_mutant/1'>See FEN1 bound to PCNA mutant</scene>. | A disease causing variant in human, which causes a large conformational change of the PIP binding pocket. While the overal structure of the S228I mutant is mostly similar to the wild type, the change causes displacements extending from the mutation site, which affect the IDCL. Notably Tyr133, a highly conservated residue, rotates outwards by nearly 90° to prevent spherical conflicts with Ile228. The changes cause the binding cavity of S228I to be about a third of the size of its wild type equivalent and makes it incompetent for PIP binding. Such changes, if they were permanent, would however be lethal, as the removal of PCNA-FEN1 interactions is lethal on its own <ref>DOI:10.1158/0008-5472.CAN-08-0168</ref>. While some client proteins (e.q. [[p21<sup>CIP1</sup>]]) are left relatively unaffected even in this state, others' binding energetics are hugely disrupted (e.q. [[RNase H2B]] or [[FEN1]]). The explaination for survival of an individual carrying this mutation is sufficient malleability of IDCL (and thus of PCNA itself). Thus a PIP might be able to initiate an "induced-fit". Even here the mutant is disadvantaged as the wild type has B-factors ~40% higher than the mutant. Thus the mutation negatively affects even the IDCL dynamics. <ref>PMID:26688547</ref>. <scene name='84/842997/Fen1_with_pcna_mutant/1'>See FEN1 bound to PCNA mutant</scene>. | ||
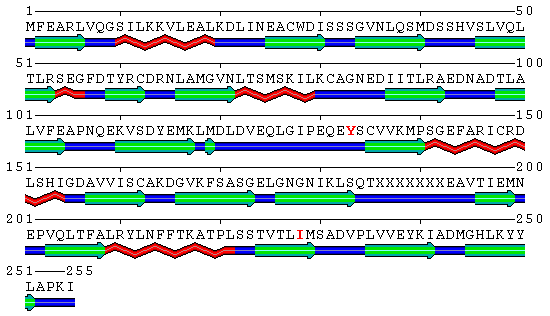
| - | [[Image:SecondaryMutantStructure.gif|frame|none|Human PCNA S228I mutant secondary structure with marked mutation and rotated Tyr133 marked. Helices are marked red, sheets green and loops blue. Note the IDCL region (residues around 125).]] | + | [[Image:SecondaryMutantStructure.gif|frame|none|Human PCNA S228I mutant secondary structure with marked mutation and rotated Tyr133 marked red in sequence. Helices are marked red, sheets green and loops blue. Note the IDCL region (residues around 125).]] |
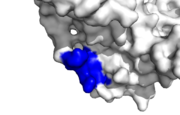
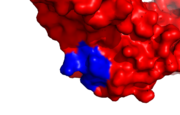
[[Image:Healthy_binding_site.png|thumb|Human PCNA PIP healthy binding site, IDCL marked blue]] | [[Image:Healthy_binding_site.png|thumb|Human PCNA PIP healthy binding site, IDCL marked blue]] | ||
Revision as of 13:13, 25 May 2020
| |||||||||||
References
- ↑ Duffy CM, Hilbert BJ, Kelch BA. A Disease-Causing Variant in PCNA Disrupts a Promiscuous Protein Binding Site. J Mol Biol. 2016 Mar 27;428(6):1023-40. doi: 10.1016/j.jmb.2015.11.029. Epub 2015, Dec 11. PMID:26688547 doi:http://dx.doi.org/10.1016/j.jmb.2015.11.029
- ↑ Fay PJ, Johanson KO, McHenry CS, Bambara RA. Size classes of products synthesized processively by two subassemblies of Escherichia coli DNA polymerase III holoenzyme. J Biol Chem. 1982 May 25;257(10):5692-9. PMID:7040370
- ↑ Yao NY, Georgescu RE, Finkelstein J, O'Donnell ME. Single-molecule analysis reveals that the lagging strand increases replisome processivity but slows replication fork progression. Proc Natl Acad Sci U S A. 2009 Aug 11;106(32):13236-41. doi:, 10.1073/pnas.0906157106. Epub 2009 Aug 3. PMID:19666586 doi:http://dx.doi.org/10.1073/pnas.0906157106
- ↑ McInerney P, Johnson A, Katz F, O'Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007 Aug 17;27(4):527-38. doi: 10.1016/j.molcel.2007.06.019. PMID:17707226 doi:http://dx.doi.org/10.1016/j.molcel.2007.06.019
- ↑ Matsumoto Y, Kim K, Hurwitz J, Gary R, Levin DS, Tomkinson AE, Park MS. Reconstitution of proliferating cell nuclear antigen-dependent repair of apurinic/apyrimidinic sites with purified human proteins. J Biol Chem. 1999 Nov 19;274(47):33703-8. doi: 10.1074/jbc.274.47.33703. PMID:10559261 doi:http://dx.doi.org/10.1074/jbc.274.47.33703
- ↑ Lee SH, Hurwitz J. Mechanism of elongation of primed DNA by DNA polymerase delta, proliferating cell nuclear antigen, and activator 1. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5672-6. doi: 10.1073/pnas.87.15.5672. PMID:1974050 doi:http://dx.doi.org/10.1073/pnas.87.15.5672
- ↑ Sanchez R, Pantoja-Uceda D, Prieto J, Diercks T, Marcaida MJ, Montoya G, Campos-Olivas R, Blanco FJ. Solution structure of human growth arrest and DNA damage 45alpha (Gadd45alpha) and its interactions with proliferating cell nuclear antigen (PCNA) and Aurora A kinase. J Biol Chem. 2010 Jul 16;285(29):22196-201. Epub 2010 May 11. PMID:20460379 doi:10.1074/jbc.M109.069344
- ↑ Kullmann F, Fadaie M, Gross V, Knuchel R, Bocker T, Steinbach P, Scholmerich J, Ruschoff J. Expression of proliferating cell nuclear antigen (PCNA) and Ki-67 in dysplasia in inflammatory bowel disease. Eur J Gastroenterol Hepatol. 1996 Apr;8(4):371-9. PMID:8781908
- ↑ De Biasio A, de Opakua AI, Mortuza GB, Molina R, Cordeiro TN, Castillo F, Villate M, Merino N, Delgado S, Gil-Carton D, Luque I, Diercks T, Bernado P, Montoya G, Blanco FJ. Structure of p15(PAF)-PCNA complex and implications for clamp sliding during DNA replication and repair. Nat Commun. 2015 Mar 12;6:6439. doi: 10.1038/ncomms7439. PMID:25762514 doi:http://dx.doi.org/10.1038/ncomms7439
- ↑ doi: https://dx.doi.org/10.1016/S0092-8674(00)81347-1
- ↑ Bruning JB, Shamoo Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1. Structure. 2004 Dec;12(12):2209-19. PMID:15576034 doi:http://dx.doi.org/10.1016/j.str.2004.09.018
- ↑ Larsen E, Kleppa L, Meza TJ, Meza-Zepeda LA, Rada C, Castellanos CG, Lien GF, Nesse GJ, Neuberger MS, Laerdahl JK, William Doughty R, Klungland A. Early-onset lymphoma and extensive embryonic apoptosis in two domain-specific Fen1 mice mutants. Cancer Res. 2008 Jun 15;68(12):4571-9. doi: 10.1158/0008-5472.CAN-08-0168. PMID:18559501 doi:http://dx.doi.org/10.1158/0008-5472.CAN-08-0168
- ↑ Duffy CM, Hilbert BJ, Kelch BA. A Disease-Causing Variant in PCNA Disrupts a Promiscuous Protein Binding Site. J Mol Biol. 2016 Mar 27;428(6):1023-40. doi: 10.1016/j.jmb.2015.11.029. Epub 2015, Dec 11. PMID:26688547 doi:http://dx.doi.org/10.1016/j.jmb.2015.11.029
- ↑ Duffy CM, Hilbert BJ, Kelch BA. A Disease-Causing Variant in PCNA Disrupts a Promiscuous Protein Binding Site. J Mol Biol. 2016 Mar 27;428(6):1023-40. doi: 10.1016/j.jmb.2015.11.029. Epub 2015, Dec 11. PMID:26688547 doi:http://dx.doi.org/10.1016/j.jmb.2015.11.029
- ↑ Lau PJ, Flores-Rozas H, Kolodner RD. Isolation and characterization of new proliferating cell nuclear antigen (POL30) mutator mutants that are defective in DNA mismatch repair. Mol Cell Biol. 2002 Oct;22(19):6669-80. doi: 10.1128/mcb.22.19.6669-6680.2002. PMID:12215524 doi:http://dx.doi.org/10.1128/mcb.22.19.6669-6680.2002
- ↑ Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, Devenney PS, Sexton D, Grimes G, Holt IJ, Hill RE, Taylor MS, Lawson KA, Dorin JR, Jackson AP. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012 May 25;149(5):1008-22. doi: 10.1016/j.cell.2012.04.011. Epub 2012 May , 10. PMID:22579044 doi:http://dx.doi.org/10.1016/j.cell.2012.04.011