We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Dora Bonadio/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
== An attractive drug target == | == An attractive drug target == | ||
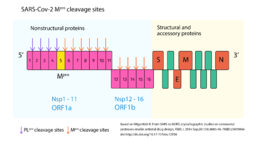
| - | As have been shown, because of its importance for viral replication, inhibiting SARS-CoV-2 Mpro activity could lead to viral replication blockage <ref name="Crystal_structure" /><ref name="ofmpro" />. Moreover, no human proteases has been reported to have a similar cleavage specificity and so, in this aspect, Mpro inhibitors toxic side-effects may be reduced <ref name="sars_mers" />. Therefore, CoV Mpro has been an attractive drug target among coronaviruses <ref name="sars_mers" /> and so it is for COVID-19 <ref name="Crystal_structure" /><ref name="ofmpro" />. Indeed, virtual drug screening, structure-assisted drug design, and high-throughput screening are been used to repurpose approved pharmaceutical drug and drug candidates targeting SARS-CoV-2 Mpro <ref name="ofmpro" /><ref name="elucidation"> Mirza, Muhammad Usman, and Matheus Froeyen. 2020. ‘Structural Elucidation of SARS-CoV-2 Vital Proteins: Computational Methods Reveal Potential Drug Candidates against Main Protease, Nsp12 Polymerase and Nsp13 Helicase’. Journal of Pharmaceutical Analysis, April. Doi:[https://doi.org/10.1016/j.jpha.2020.04.008].</ref>. Furthermore, a study carrying the pharmacokinetic characterization of an optimized Mpro <scene name='84/845941/13b/1'>α-ketoamide inhibitor</scene> provides useful framework for development of this kind of inhibitors toward coronaviruses <ref name="Crystal_structure" />. It was showed that the α-ketoamide inhibitor interacts with the catalytic site of the enzyme through two hydrogen bonding interactions, as can be seen in the complex formed between SARS-CoV-2 Mpro and an α-ketoamide inhibitor <ref name="Crystal_structure" />. | + | As have been shown, because of its importance for viral replication, inhibiting SARS-CoV-2 Mpro activity could lead to viral replication blockage <ref name="Crystal_structure" /><ref name="ofmpro" />. Moreover, no human proteases has been reported to have a similar cleavage specificity and so, in this aspect, Mpro inhibitors toxic side-effects may be reduced <ref name="sars_mers" />. Therefore, CoV Mpro has been an attractive drug target among coronaviruses <ref name="sars_mers" /> and so it is for COVID-19 <ref name="Crystal_structure" /><ref name="ofmpro" />. Indeed, virtual drug screening, structure-assisted drug design, and high-throughput screening are been used to repurpose approved pharmaceutical drug and drug candidates targeting SARS-CoV-2 Mpro <ref name="ofmpro" /><ref name="elucidation"> Mirza, Muhammad Usman, and Matheus Froeyen. 2020. ‘Structural Elucidation of SARS-CoV-2 Vital Proteins: Computational Methods Reveal Potential Drug Candidates against Main Protease, Nsp12 Polymerase and Nsp13 Helicase’. Journal of Pharmaceutical Analysis, April. Doi:[https://doi.org/10.1016/j.jpha.2020.04.008].</ref>. Furthermore, a study carrying the pharmacokinetic characterization of an optimized Mpro <scene name='84/845941/13b/1'>α-ketoamide inhibitor</scene> provides useful framework for development of this kind of inhibitors toward coronaviruses <ref name="Crystal_structure" />. It was showed that the α-ketoamide inhibitor interacts with the catalytic site of the enzyme through two hydrogen bonding interactions, as can be seen in the complex formed between SARS-CoV-2 Mpro and an <scene name='84/845941/13b2/1'>α-ketoamide inhibitor</scene> <ref name="Crystal_structure" />. |
== External Resources == | == External Resources == | ||
Revision as of 08:13, 11 June 2020
SARS-CoV-2 main protease (Mpro)
| |||||||||||