User:Felipe de Melo Santana/Sandbox 1
From Proteopedia
| Line 10: | Line 10: | ||
== Structure == | == Structure == | ||
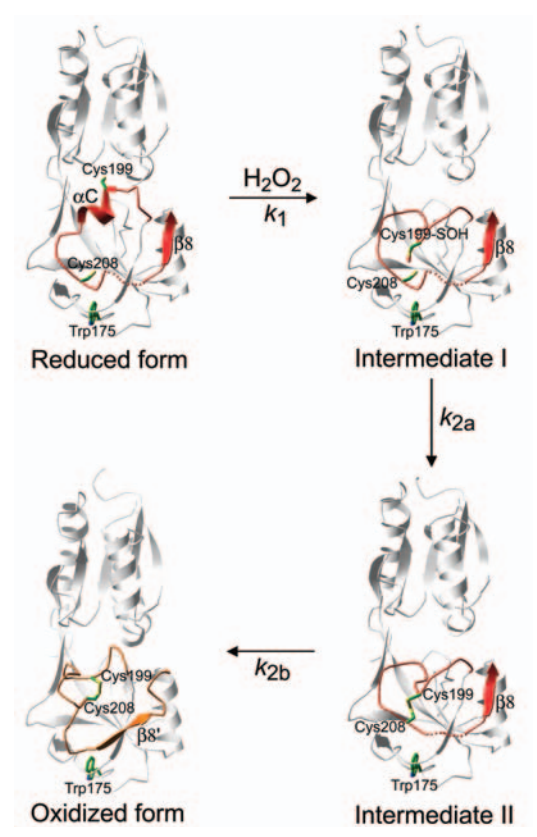
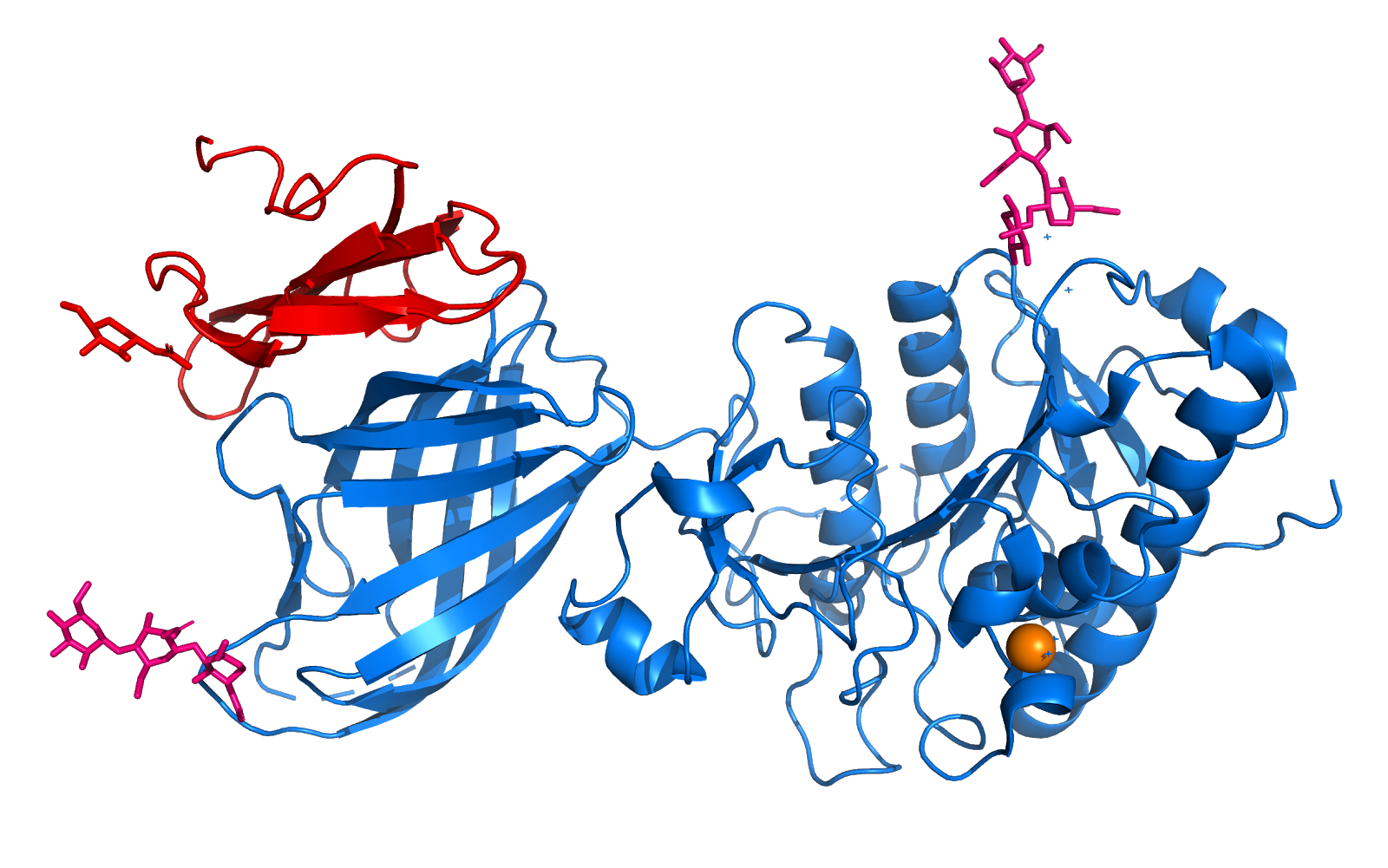
| - | This protein its a transcription factor, at that time its shows two different structures when its reduced or oxidized. The reduced form monomer of the OxyR regulatory domain consists of two α/β domains that are linked by two interdomain strands. Residues <scene name='84/844928/87-162/1'>87–162</scene> in the middle of a folded domain form the majority of the N-terminal a/b domain. The C-terminal a/b Results and Discussion domain II is made of residues 164–260. | + | This protein its a transcription factor, at that time its shows two different structures when its reduced or oxidized. The reduced form monomer of the OxyR regulatory domain consists of two α/β domains that are linked by two interdomain strands. Residues <scene name='84/844928/87-162/1'>87–162</scene> in the middle of a folded domain form the majority of the N-terminal a/b domain. The C-terminal a/b Results and Discussion domain II is made of residues <scene name='84/844928/164-260/1'>164–260</scene>. |
The remaining C-terminal 30 Structure Determination residues come back to domain I to form an edge of the domain . The two domains exhibit a similar folding pattern consisting of a central b sheet flanked by helices and loops. | The remaining C-terminal 30 Structure Determination residues come back to domain I to form an edge of the domain . The two domains exhibit a similar folding pattern consisting of a central b sheet flanked by helices and loops. | ||
Revision as of 01:35, 17 June 2020
1I69
| |||||||||||
References
Zheng, M., Aslund, F., and Storz, G. (1998). Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279, 1718–1721.
Choi H, Kim S, Mukhopadhyay P, et al. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105(1):103-113. doi:10.1016/s0092-8674(01)00300-2
Toledano, M.B., Kullik, I., Trinh, F., Baird, P.T., Schneider, T.D., and Storz, G. (1994). Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78, 897–909.
Storz, G., Tartaglia, L.A., and Ames, B.N. (1990). Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248, 189–194
Kullik, I., Toledano, M.B., Tartaglia, L.A., and Storz, G. (1995a). Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J. Bacteriol. 177, 1275–1284.
Lee C, Lee SM, Mukhopadhyay P, et al. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat Struct Mol Biol. 2004;11(12):1179-1185. doi:10.1038/nsmb856
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644