User:Felipe de Melo Santana/Sandbox 1
From Proteopedia
| Line 22: | Line 22: | ||
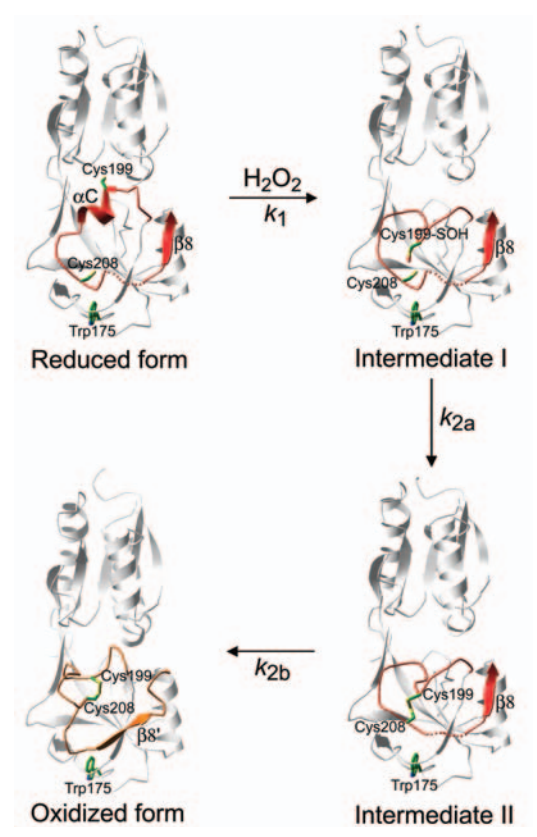
The OxyR activation, by H2O2, begins with the oxidation of the <scene name='84/844928/Cys_199/1'>Cys199</scene> residue into a sulfenic acid intermediate, which causes destabilization of the Cys199 side chain in the hydrophobic binding pocket, due to its increased size and charge. The destabilization leads to expulsion of the side chain of Cys199 out of the interdomain pocket, resulting in a flexible loop around Cys199 (Lee et al. 2004). | The OxyR activation, by H2O2, begins with the oxidation of the <scene name='84/844928/Cys_199/1'>Cys199</scene> residue into a sulfenic acid intermediate, which causes destabilization of the Cys199 side chain in the hydrophobic binding pocket, due to its increased size and charge. The destabilization leads to expulsion of the side chain of Cys199 out of the interdomain pocket, resulting in a flexible loop around Cys199 (Lee et al. 2004). | ||
| - | This change in the regulatory domain of OxyR allows the formation of an intramolecular disulfide bond between Cys199 and Cys208, responsible for keeping OxyR in the active form. Thus, OxyR is deactivated by the reduction of the disulfide bond, that leads OxyR back to its reduced form (Zheng et al. 1998). | + | This change in the regulatory domain of OxyR allows the formation of an intramolecular disulfide bond between <scene name='84/844928/Cys_199_-_cys_208/1'>Cys199 and Cys208</scene>, responsible for keeping OxyR in the active form. Thus, OxyR is deactivated by the reduction of the disulfide bond, that leads OxyR back to its reduced form (Zheng et al. 1998). |
Revision as of 19:46, 20 June 2020
1I69
| |||||||||||
References
Zheng, M., Aslund, F., and Storz, G. (1998). Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279, 1718–1721.
Choi H, Kim S, Mukhopadhyay P, et al. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105(1):103-113. doi:10.1016/s0092-8674(01)00300-2
Toledano, M.B., Kullik, I., Trinh, F., Baird, P.T., Schneider, T.D., and Storz, G. (1994). Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78, 897–909.
Storz, G., Tartaglia, L.A., and Ames, B.N. (1990). Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248, 189–194
Kullik, I., Toledano, M.B., Tartaglia, L.A., and Storz, G. (1995a). Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J. Bacteriol. 177, 1275–1284.
Lee C, Lee SM, Mukhopadhyay P, et al. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat Struct Mol Biol. 2004;11(12):1179-1185. doi:10.1038/nsmb856
Previato, M. Regulation of genes involved in oxidative stress response in Caulobacter crescentus. 2013. 134 p. Masters thesis (Microbiology) – Instituto de Ciências Biomédicas, Universidade de São Paulo, São Paulo, 2013.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644