We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Andre Wu Le Chun/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
== Structural highlights == | == Structural highlights == | ||
| - | <scene name='84/844931/6vsb/2'>6vsb</scene> is a homotrimer transmembrane glycoprotein(S) protruding from the viral surface¹. It is composed by two subunits that are fundamental for the virus entry in the cell: The S1 subunit, which contains the receptor binding domain (<scene name='84/844931/S_receptor-binding_domain/1'>RBD</scene>), is responsible for binding to the host cell receptor, and the S2 subunit, responsible for fusion of the viral and cellular membranes. In order to allow the binding of the RBD in S1 with the host cell, the spike protein goes through conformational changes that make the binding domain assume a up conformation, thus, exposing the spike protein to its target receptor. | + | <scene name='84/844931/6vsb/2'>6vsb</scene> is a homotrimer transmembrane glycoprotein(S) protruding from the viral surface¹. It is composed by two subunits that are fundamental for the virus entry in the cell: The S1 subunit, a v shaped subunit which contains the receptor binding domain (<scene name='84/844931/S_receptor-binding_domain/1'>RBD</scene>), is responsible for binding to the host cell receptor, and the S2 subunit, responsible for fusion of the viral and cellular membranes. In order to allow the binding of the RBD in S1 with the host cell, the spike protein goes through conformational changes that make the binding domain assume a up conformation, thus, exposing the spike protein to its target receptor. |
| Line 23: | Line 23: | ||
== Disease == | == Disease == | ||
6vsb is probably the protein that enabled the Sars-Cov-2 to enter human cells, rusulting therefore on the large scale coronavirus epidemics in 2020. | 6vsb is probably the protein that enabled the Sars-Cov-2 to enter human cells, rusulting therefore on the large scale coronavirus epidemics in 2020. | ||
| - | + | The main symptoms of the COVID-19 were similar to the ones of common cold: fever, cough and tiredness, but with some stronger than the common cold: Chest pain, lost of smell and taste sense. | |
| - | The main reason for the worst symptoms | + | The main reason for the worst symptoms is the death of several types of cells in the lungs, causing severe loss of oxygenation, therefore, resulting of gradual colapse of the respiratory system of the infected. |
== Relevance == | == Relevance == | ||
| Line 30: | Line 30: | ||
== Interaction with angiotensin-converting enzime 2 == | == Interaction with angiotensin-converting enzime 2 == | ||
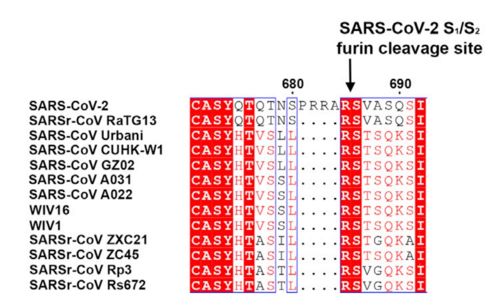
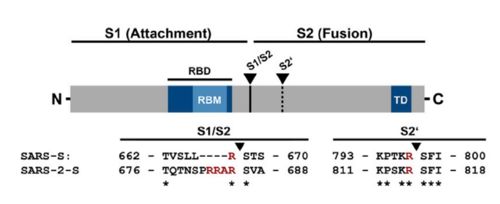
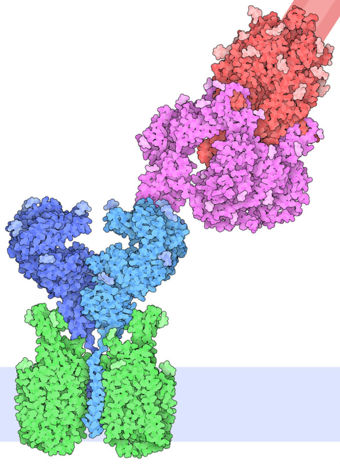
| - | The interaction between the 2019-nCov and the host cell begins with the recognition of the ACE2, a dimeric protein. Then, the S1 subunit moves, modifying the protein's conformation in way that receptor-binding domain becomes available to connect itself to the peptidase domain of | + | The interaction between the 2019-nCov and the host cell begins with the recognition of the ACE2, a dimeric protein. Then, the S1 subunit moves, modifying the protein's conformation in way that receptor-binding domain becomes available to connect itself to the peptidase domain of the receptor. At that instance, the spike protein is found in a "up" conformation. Due to the existence of four amino acids in between the 680 and 685 positions, a furin cleavage site is featured in the protein. Once this site is cleaved by host's furins, a later cleavage at S2' by a serine protease, TMPRSS2, happens in order to allow the activation of the fusion mechanism. Lastly, the spike protein makes a high affinity bond with ACE2 at the order of 15nM and a complex of spike protein and ACE2, as shown in figure 3, is formed. |
[[Image:Spike.jpg|500px|]] | [[Image:Spike.jpg|500px|]] | ||
Figure 2: Alignment between S protein of SARS and SARS-2 (Hoffmann et al., 2020). | Figure 2: Alignment between S protein of SARS and SARS-2 (Hoffmann et al., 2020). | ||
[[Image:SpikeACE2.jpg|340px|]] | [[Image:SpikeACE2.jpg|340px|]] | ||
| - | Figure 3: Spike protein and ACE2 bound together ( | + | Figure 3: Spike protein and ACE2 bound together (Goodsell, D., 2020) |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
Revision as of 21:41, 21 June 2020
6vsb
Prefusion 2019-nCoV spike glycoprotein with a single receptor-binding domain up
| |||||||||||
References
¹https://www.sciencedirect.com/science/article/pii/S0065352719300284 ²https://www.biorxiv.org/content/10.1101/2020.02.19.956581v1.full https://www.sciencedirect.com/science/article/pii/S0092867420302294
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644