User:Andre Wu Le Chun/Sandbox 1
From Proteopedia
| Line 6: | Line 6: | ||
== Structural highlights == | == Structural highlights == | ||
| - | <scene name='84/844931/6vsb/2'>6vsb</scene> is a homotrimer transmembrane glycoprotein(S) protruding from the viral surface. It is composed by two subunits that are fundamental for the virus entry in the cell: The S1 subunit, a v shaped subunit which contains the receptor binding domain (<scene name='84/844931/S_receptor-binding_domain/1'>RBD</scene>), is responsible for binding to the host cell receptor, and the S2 subunit, which is responsible for fusion of the viral and cellular membranes. In order to allow the binding of the RBD in S1 with the host cell, the spike protein goes through conformational changes that make the binding domain assume a up conformation, thus, exposing the spike protein to its target receptor. | + | <scene name='84/844931/6vsb/2'>6vsb</scene> is a homotrimer transmembrane glycoprotein(S) protruding from the viral surface. It is composed by two subunits that are fundamental for the virus entry in the cell: The S1 subunit, a v shaped subunit which contains the receptor binding domain (<scene name='84/844931/S_receptor-binding_domain/1'>RBD</scene>), which consists of the residues Arg319-Phe541, is responsible for binding to the host cell receptor, and the S2 subunit, which is responsible for fusion of the viral and cellular membranes. In order to allow the binding of the RBD in S1 with the host cell, the spike protein goes through conformational changes that make the binding domain assume a up conformation, thus, exposing the spike protein to its target receptor. |
| Line 29: | Line 29: | ||
== Interaction with angiotensin-converting enzime 2 == | == Interaction with angiotensin-converting enzime 2 == | ||
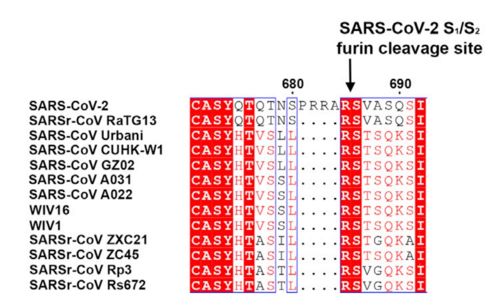
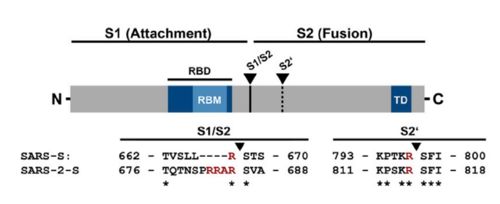
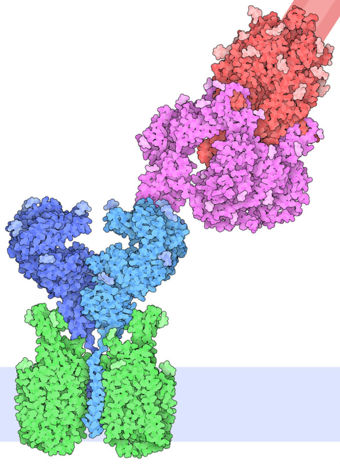
| - | The interaction between the 2019-nCov and the host cell begins with the recognition of the ACE2, a dimeric protein. Then, the S1 subunit moves, modifying the protein's conformation in way that receptor-binding domain becomes available to connect itself to the peptidase domain of the receptor. At that moment, the spike protein is found in a "up" conformation. Due to the existence of four amino acids in between the 680 and 685 positions, a furin cleavage site is featured in the protein. Once this site is cleaved by host's furins, a later cleavage at S2' by a serine protease, TMPRSS2, happens in order to allow the activation of the fusion mechanism. Lastly, the spike protein makes a high affinity bond with ACE2 at the order of 15nM and a complex of spike protein and ACE2, as shown in figure 3, is formed. In order for the biding to happen 17 amino acid residues of the RBD interact with 20 amino acid residues of the receptor ACE2. | + | The interaction between the 2019-nCov and the host cell begins with the recognition of the ACE2, a dimeric protein. Then, the S1 subunit moves, modifying the protein's conformation in way that receptor-binding domain becomes available to connect itself to the peptidase domain of the receptor. At that moment, the spike protein is found in a "up" conformation. Due to the existence of four amino acids in between the 680 and 685 positions, a furin cleavage site is featured in the protein. Once this site is cleaved by host's furins, a later cleavage at S2' by a serine protease, TMPRSS2, happens in order to allow the activation of the fusion mechanism. Lastly, the spike protein makes a high affinity bond with ACE2 at the order of 15nM and a complex of spike protein and ACE2, as shown in figure 3, is formed. In order for the biding to happen 17 amino acid residues of the RBD interact with 20 amino acid residues of the receptor ACE2. It is important to consider the receptor binding domain similarity of both the SARS-CoV and the SARS-CoV-2. 14 amino acid positions are shared between the two conoraviruses RBD, these include Tyr449/Tyr436, Tyr453/Tyr440, Asn487/Asn473, Tyr489/Tyr475, Gly496/Gly482, Thr500/Thr486, Gly502/Gly488 and Tyr505/Tyr491, which are identical, and other 6 amino acid positions with altered residues. Also in the RBD hydrophilic interctions such as 13 hydrogen bonds and 3 salt bridges can be found |
[[Image:Spike.jpg|500px|]] | [[Image:Spike.jpg|500px|]] | ||
Revision as of 20:44, 31 July 2020
6vsb
Prefusion 2019-nCoV spike glycoprotein with a single receptor-binding domain up
| |||||||||||
Hoffmann, Markus & Kleine-Weber, Hannah & Schroeder, Simon & Krüger, Nadine & Herrler, Tanja & Erichsen, Sandra & Schiergens, Tobias & Herrler, Georg & Wu, Nai-Huei & Nitsche, Andreas & Müller, Marcel & Drosten, Christian &
Pöhlmann, Stefan. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 181. 10.1016/j.cell.2020.02.052.
Tortorici, M. Alejandra & Veesler, David. (2019). Structural insights into coronavirus entry. 10.1016/bs.aivir.2019.08.002.
Walls, Alexandra & Park, Young-Jun & Tortorici, M. & Wall, Abigail & Mcguire, Andrew & Veesler, David. (2020). Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein. 10.1101/2020.02.19.956581.