Talk:Hen Egg-White (HEW) Lysozyme
From Proteopedia
(→3D scenes) |
|||
| Line 61: | Line 61: | ||
<scene name='37/376372/Secondary_structure/7'>37/376372/Secondary_structure/7</scene> | <scene name='37/376372/Secondary_structure/7'>37/376372/Secondary_structure/7</scene> | ||
| + | |||
| + | <scene name='37/376372/Hbonds/1'>37/376372/Hbonds/1</scene> | ||
| + | |||
| + | <scene name='37/376372/Hbonds/5'>37/376372/Hbonds/5</scene> | ||
| + | |||
| + | <scene name='37/376372/Hbonds/2'>37/376372/Hbonds/2</scene> | ||
| + | |||
| + | <scene name='37/376372/Hbonds/3'>37/376372/Hbonds/3</scene> | ||
| + | |||
| + | <scene name='37/376372/Hex_model/1'>37/376372/Hex_model/1</scene> | ||
| + | |||
| + | <scene name='37/376372/Covalent_intermediate/2'>37/376372/Covalent_intermediate/2</scene> | ||
| + | |||
| + | <scene name='37/376372/Product_complex/6'>37/376372/Product_complex/6</scene> | ||
| + | |||
| + | |||
| + | <scene name=''></scene> | ||
| + | |||
| + | |||
| + | <scene name=''></scene> | ||
| + | |||
| + | |||
| + | <scene name=''></scene> | ||
| + | |||
| + | |||
| + | <scene name=''></scene> | ||
| + | |||
| + | |||
| + | <scene name=''></scene> | ||
| + | |||
| + | <scene name=''></scene> | ||
| - | Anything in this section will appear adjacent to the 3D structure and will be scrollable. | ||
</StructureSection> | </StructureSection> | ||
Revision as of 12:22, 2 August 2020
How some of the figures were made
Opening scene
This is a script to create the opening scene after loading coordinate 1HEW (you could also use the screen authoring tool)
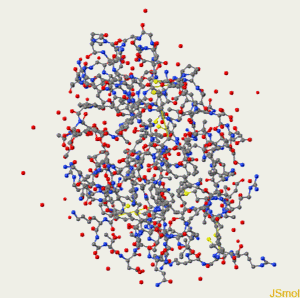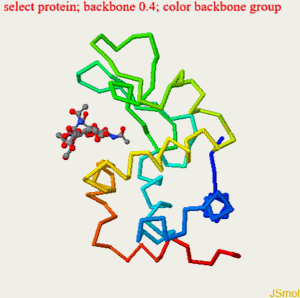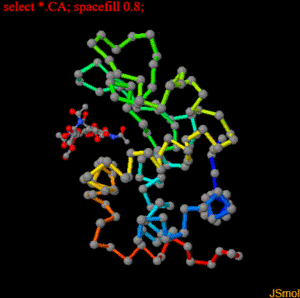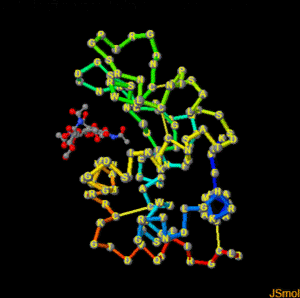
restrict none #clean slate select ligand; wireframe 0.3; spacefill 0.5; #make bonds on ligand a bit thicker select protein; backbone 0.4; color backbone group #rainbow c-alpha trace color background black #as it says select *.CA; spacefill 0.8; #spheres on alpha carbons label "%m";set labelOffset 0 0; color label yellow #...with yellow labels on top hover "%n %r, Chain=%c, Element=%e"; #show 3-letter code number, chain and element select cys.sg; wireframe 0.25; set ssbonds backbone #disulfide bonds
Here is an animation to see what the commands do
and a screen shot of the finished scene with captions for reference
Hydrogen bonds
To enable centering with the mouse for this scene ("set picking center"), the green link was made as a JmolLink rather than using the Screen Authoring tool. The following was used:
<jmol>
<jmolLink>
<script>set picking center; display all; script /scripts/37/376372/Hbonds/5.spt </script>
<text>hydrogen bonds</text>
</jmolLink>
</jmol>
Except for the two Jmol commands that are explicitly shown above, the rest of the figure was saved as the usual state file in the scene authoring tool, and then called to generate the figure ("script ...Hbonds/5.spt"). For the other green links in the section, I was able to use the the conventional method because the command "set picking center" had already been issued. Below are the commands to show the hydrogen bonds in the context of the protein, ligand and water.
restrict none # clean slate select protein; wireframe 0.1 # thin bonds for protein select backbone; wireframe 0.3 # but thick bonds for main chain set ssbondsbackbone false # disulfide bonds between sulfur atoms, not C-alpha color background white # as it says select water; spacefill 0.2; # water oxygens as spheres select ligand; spacefill 0.6; wireframe 0.3 # ligand in ball and stick select ligand and _C;color atoms opaque [x800080] # carbon atoms of ligand in purple set hbondsRasmol FALSE; calculate HBONDS # calculate and display hydrogen bonds (not just main chain) slab 60;depth 20;slab on # slab away atoms in foreground set zShade true # fade away atoms in background
3D scenes
| |||||||||||
Lysozyme list of figures
--Karsten Theis 01:42, 20 July 2019 (UTC)
Initial view
script: https://proteopedia.org/wiki/scripts/37/376372/Overall/3.spt
coords: https://proteopedia.org/cgi-bin/getfrozenstructure?2f267243b1cf263a27c03cd5ce051b66
Overview of the 1HEW structure. The trace of the protein chain is shown in rainbow colors with the alpha carbons as gray spheres labeled with the residue one letter code. The carbohydrate ligand is shown as ball and stick in CPK colors (carbon: gray, oxygen: red, nitrogen: blue).
ribbon view
script: https://proteopedia.org/wiki/scripts/37/376372/Secondary_structure/7.spt
coords: same as Initial view
Overview of the 1HEW secondary structure: Alpha Helices, 3-10 helix, Beta Strands, with loops in white and disulfide bonds in yellow. Arrow tips point towards the C-terminal end.
colored by conservation
script: https://proteopedia.org/wiki/ConSurf/he/1hew_consurf.spt coords: none
helices
script: https://proteopedia.org/wiki/scripts/37/376372/Hbonds/1.spt
coords: same as Initial view
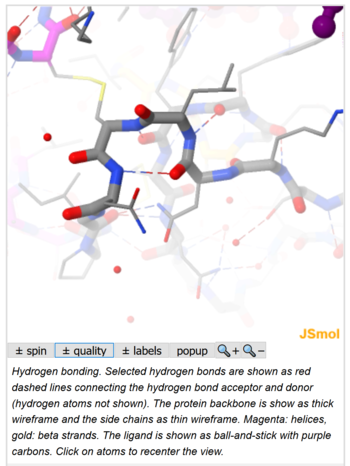
Hydrogen bonding. Selected hydrogen bonds are shown as red dashed lines connecting the hydrogen bond acceptor and donor (hydrogen atoms not shown). The protein backbone is show as thick wireframe and the side chains as thin wireframe. Magenta: helices, gold: beta strands. The ligand is shown as ball-and-stick with purple carbons. Click on atoms to recenter the view.
beta turns
script: https://proteopedia.org/wiki/scripts/37/376372/Hbonds/5.spt
coords: same as Initial view
Hydrogen bonding. Selected hydrogen bonds are shown as red dashed lines connecting the hydrogen bond acceptor and donor (hydrogen atoms not shown). The protein backbone is show as thick wireframe and the side chains as thin wireframe. Magenta: helices, gold: beta strands. The ligand is shown as ball-and-stick with purple carbons. Click on atoms to recenter the view.
between strands
script: https://proteopedia.org/wiki/scripts/37/376372/Hbonds/2.spt
coords: same as Initial view
Hydrogen bonding. Selected hydrogen bonds are shown as red dashed lines connecting the hydrogen bond acceptor and donor (hydrogen atoms not shown). The protein backbone is show as thick wireframe and the side chains as thin wireframe. Magenta: helices, gold: beta strands. The ligand is shown as ball-and-stick with purple carbons. Click on atoms to recenter the view.
between protein and ligand
script: https://proteopedia.org/wiki/scripts/37/376372/Hbonds/3.spt
coords: same as Initial view
Hydrogen bonding. Selected hydrogen bonds are shown as red dashed lines connecting the hydrogen bond acceptor and donor (hydrogen atoms not shown). The protein backbone is show as thick wireframe and the side chains as thin wireframe. Magenta: helices, gold: beta strands. The ligand is shown as ball-and-stick with purple carbons. Click on atoms to recenter the view.
this hypothetical model
script: https://proteopedia.org/wiki/scripts/37/376372/Hex_model/1.spt
coords: https://proteopedia.org/cgi-bin/getfrozenstructure?caa67c01c8140e2b3f7b60e4e749f1dd
1HEW with additional three sugars modeled into the active site.
covalent intermediate
script: https://proteopedia.org/wiki/scripts/37/376372/Covalent_intermediate/2.spt
coords: https://proteopedia.org/cgi-bin/getfrozenstructure?932c10afc4e1e94cdeab12d300cbb47b (always reloads)
Structure of the covalent enzyme intermediate. The active site residue Asp 52 and the carbohydrate are shown in CPK colors, with the 2FO-FC electron density within 1.5 A of these atoms as blue mesh. The backbone trace is shown in gold.
product complex with five sugar units
script: https://proteopedia.org/wiki/scripts/37/376372/Product_complex/6.spt
coords: https://proteopedia.org/cgi-bin/getfrozenstructure?09a5df19589b9b4571d13eef460f52bf
Comparison of covalent intermediate (1H6M) and product complex (2WAR), both for the almost inactive Glu35Gln mutant. Backbone in gold, with ligand in thick wireframe and side chains of active site residues 35 and 52 in thin wireframe. Fluorine is shown in green.
Jmol commands: buttons etc
Lysozyme
script /scripts/37/376372/Overall/3.spt; hide water; set zshade off
☼
select ligand; selectionHalos ON; delay 0.5;selectionHalos OFF;
☼
select 1.CA; selectionHalos ON; delay 0.5;selectionHalos OFF;
☼
select 129.CA; selectionHalos ON; delay 0.5;selectionHalos OFF;
☼
select BONDS ({0:3}); wireframe 0.6; delay 0.5; wireframe 0.1
☼
select helix and *.CA; selectionHalos ON; delay 0.5;selectionHalos OFF;
☼
select sheet and *.CA; selectionHalos ON; delay 0.5;selectionHalos OFF;
Hydrophobic residues
select backbone or water; spacefill off; hide ligand or (sidechain and not (met, cys, ile, leu, val, phe, tyr, trp))
hydrophilic residues and proline
select backbone or water; spacefill off; hide ligand or (sidechain and (met, cys, ile, leu, val, phe, tyr, trp))
water
select backbone; spacefill on; select water; spacefill 1.0; display all
Choice of
- hydrophobic side chains: hide (hidden and not sidechain) or (sidechain and not (met, cys, ile, leu, val, phe, tyr, trp))
- hydrophilic side chains: hide (hidden and not sidechain) or (sidechain and (met, cys, ile, leu, val, phe, tyr, trp))
- all side chains: display (displayed and not sidechain) or sidechain
Choice of
- colored by hydrophobiticity: background white; select sidechain; color magenta; select sidechain and (cys, met, ile, leu, val, phe, tyr, trp); color gray; set echo ID bla 80% 0%; echo \"hydrophobic\"; color echo gray; frank off; set echo ID bla2 0% 0%; echo \"hydrophilic\"; color echo magenta
- colored by charge: select sidechain; color white; select sidechain and (asp, glu); color red; select sidechain and (lys, arg, his); color blue; set echo ID bla 80% 0%; echo \"positive\"; color echo blue; frank off; set echo ID bla2 0% 0%; echo \"negative\"; color echo red
- colored by conservation: select sidechain; define ~consurf_to_do selected; define ~consurf_to_color selected; useFullScript = true; consurf_initial_scene = false; script /wiki/ConSurf/he/1hew_consurf.spt; set echo ID bla 80% 0%; echo \"conserved\"; color echo firebrick; frank off; set echo ID bla2 0% 0%; echo \"variable\"; color echo darkturquoise
[± backbone spacefill]
if ({*.O and backbone and visible}.size > 0) {select backbone; spacefill off} else {select backbone; spacefill on}
[± ligand]
if ({ligand and hidden}.size > 0) {display displayed or ligand} else {hide ligand or hidden}
[± water]
if ({water and visible}.size > 0) {select water; spacefill off} else {select water; spacefill 1.0}
hydrogen bonds
set picking center; display all; script /scripts/37/376372/Hbonds/5.spt
Ordered waters
hide none; script /scripts/37/376372/Hbonds/4.spt
toggle fading and slabbing
if (zshade) {set zshade off; slab off} else {set zshade on; slab on}
turn centering on
set picking center
turn centering off
set picking identify
modeled extra sugars
- [On]: display all
- [Off]: hide 204-206
Choice of
- product: anim off; delay 1.0; model 1
- intermediate: anim off; delay 1.0; model 2
- animate: anim fps 1; anim mode loop; anim on;
just ring
- [On]: hide ligand and not ((1134 or G2F) and (*.O1, *.C1, *.C2, *.C3, *.C4, *.C5, *.O5, *.N5, *.O4))
- [Off]: hide water
Summary
Total of green links: 10
Total of Jmol buttons etc: 23
coordinates used
- /cgi-bin/getfrozenstructure?2f267243b1cf263a27c03cd5ce051b66
- /cgi-bin/getfrozenstructure?caa67c01c8140e2b3f7b60e4e749f1dd
- /cgi-bin/getfrozenstructure?932c10afc4e1e94cdeab12d300cbb47b
- /cgi-bin/getfrozenstructure?09a5df19589b9b4571d13eef460f52bf
Substrate complex with sugar not in chair conformation
1QBB, 1OVW, discussed in https://pubs.acs.org/doi/pdfplus/10.1021/jacs.5b01156