Lysine-specific demethylase 1 (LSD-1)
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
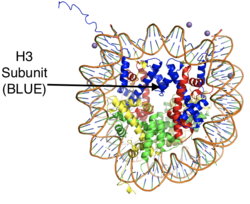
<scene name='83/834203/Overall_lsd-1/1'>LSD-1</scene>, human lysine-specific demethylase 1, is an enzyme that affects the ability of DNA to associate with [https://en.wikipedia.org/wiki/Histone histone proteins]. Histone proteins contain residues that give them an overall positive charge and allow them to act as spools upon which negatively charged DNA can wrap around for storage in the nucleus of a cell (Figure 1). When DNA is tightly condensed it forms into nucleosomes which consist of eight histone core proteins (2 H2A, 2 H2B, 2 H3, 2 H4) with DNA tightly coiled around them. This tightly coiled DNA is known as [https://en.wikipedia.org/wiki/Heterochromatin heterochromatin], which is inaccessible to transcription factors and RNA polymerase. This can be reversed by modifications to the histone protein structure that cause the DNA to relax and form [https://en.wikipedia.org/wiki/Euchromatin euchromatin], which allows for gene expression. One key histone modification is the [https://en.wikipedia.org/wiki/Methyltransferase methylation] and subsequent [https://en.wikipedia.org/wiki/Demethylase demethylation] of lysine residues. Before 2004, it was believed that methylation of histone tails was stable and irreversible. In 2004, it was discovered that histone tails can also be demethylated by demethylase enzymes such as LSD-1.<ref name="Shi">doi: 10.1016/j.cell.2004.12.012</ref> | <scene name='83/834203/Overall_lsd-1/1'>LSD-1</scene>, human lysine-specific demethylase 1, is an enzyme that affects the ability of DNA to associate with [https://en.wikipedia.org/wiki/Histone histone proteins]. Histone proteins contain residues that give them an overall positive charge and allow them to act as spools upon which negatively charged DNA can wrap around for storage in the nucleus of a cell (Figure 1). When DNA is tightly condensed it forms into nucleosomes which consist of eight histone core proteins (2 H2A, 2 H2B, 2 H3, 2 H4) with DNA tightly coiled around them. This tightly coiled DNA is known as [https://en.wikipedia.org/wiki/Heterochromatin heterochromatin], which is inaccessible to transcription factors and RNA polymerase. This can be reversed by modifications to the histone protein structure that cause the DNA to relax and form [https://en.wikipedia.org/wiki/Euchromatin euchromatin], which allows for gene expression. One key histone modification is the [https://en.wikipedia.org/wiki/Methyltransferase methylation] and subsequent [https://en.wikipedia.org/wiki/Demethylase demethylation] of lysine residues. Before 2004, it was believed that methylation of histone tails was stable and irreversible. In 2004, it was discovered that histone tails can also be demethylated by demethylase enzymes such as LSD-1.<ref name="Shi">doi: 10.1016/j.cell.2004.12.012</ref> | ||
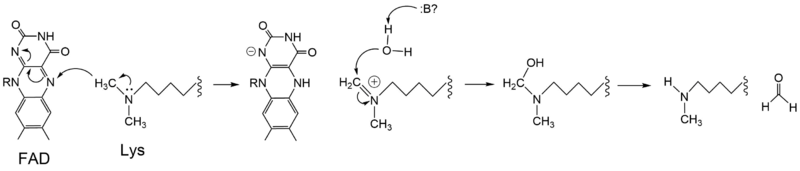
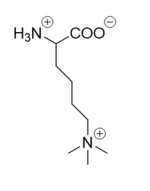
| - | There are two main classes of demethylases, and they are categorized by their co-factors and reaction mechanisms. One class of demethylases (KDM1) uses a flavin adenine dinucleotide [https://en.wikipedia.org/wiki/Flavin_adenine_dinucleotide FAD] co-factor to catalyze the demethylation reaction. The other class of demethylases (KDM2-6) uses a Fe<sup>2+</sup> ion and [https://en.wikipedia.org/wiki/Alpha-Ketoglutaric_acid α-ketoglutarate] as a co-substrate to catalyze the reaction. Although the co-factors used are different, both classes operate by hydroxylating the target methyl group. LSD-1 is in the KDM1 family of histone demthylases that uses FAD as a co-factor. LSD-1 specifically demethylates mono- or di-methylated lysine substrates at Lys4 or Lys9 in the tail of histone H3. <ref name="Forneris">PMID: 15811342</ref> Demethylation of these lysine residues is commonly associated with transcriptional activation, but it also has the ability to silence genes depending on the residue being demethylated, the cofactors present, and the environment in which the demethylation occurs. | + | There are two main classes of demethylases, and they are categorized by their co-factors and reaction mechanisms. One class of demethylases (KDM1) uses a flavin adenine dinucleotide [https://en.wikipedia.org/wiki/Flavin_adenine_dinucleotide FAD] co-factor to catalyze the demethylation reaction. The other class of demethylases (KDM2-6) uses a Fe<sup>2+</sup> ion and [https://en.wikipedia.org/wiki/Alpha-Ketoglutaric_acid α-ketoglutarate] as a co-substrate to catalyze the reaction. Although the co-factors used are different, both classes operate by hydroxylating the target methyl group. LSD-1 is in the KDM1 family of histone demthylases that uses FAD as a co-factor. LSD-1 specifically demethylates mono- or di-methylated lysine substrates at Lys4 or Lys9 in the tail of histone H3.<ref name="Forneris">PMID: 15811342</ref> Demethylation of these lysine residues is commonly associated with transcriptional activation, but it also has the ability to silence genes depending on the residue being demethylated, the cofactors present, and the environment in which the demethylation occurs. |
== Structure == | == Structure == | ||
| - | The human lysine-specific demethylase-1 structure was determined with a resolution of 2.9 Å.<ref name="Stavropolous">doi: 10.1038/nsmb1113</ref> The crystals were grown without the first 165 residues due to their proposed lack of structure and protease susceptibility. <ref name="Stavropolous"/> Additionally, two unstructured loops (residues 467–474 and 784–792) remained unresolved in the final refined structure. <ref name="Stavropolous"/> | + | The human lysine-specific demethylase-1 structure was determined with a resolution of 2.9 Å.<ref name="Stavropolous">doi: 10.1038/nsmb1113</ref> The crystals were grown without the first 165 residues due to their proposed lack of structure and protease susceptibility.<ref name="Stavropolous"/> Additionally, two unstructured loops (residues 467–474 and 784–792) remained unresolved in the final refined structure.<ref name="Stavropolous"/> |
=== Tower Domain === | === Tower Domain === | ||
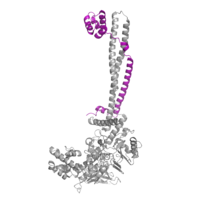
[[Image:COREST.png|200 px|left|thumb|Figure 2: CoRest complex (purple) bound to LSD-1 (PDB: 2IW5) at the Tower domain.]] | [[Image:COREST.png|200 px|left|thumb|Figure 2: CoRest complex (purple) bound to LSD-1 (PDB: 2IW5) at the Tower domain.]] | ||
| - | The <scene name='83/834203/Towerdomain/3'>tower domain</scene> is a 100 residue protrusion off of the main protein body that is caused by an insertion into the sequence of the oxidase domain. It is comprised of a coiled-coil of two [https://en.wikipedia.org/wiki/Alpha_helix α-helices]. The longer helix, TαA, is an LSD-1 specific element that has not been found in other oxidase proteins. <ref name="Stavropolous"/> The shorter helix, TαB, connects directly to helix αD of the oxidase domain through a conserved connector loop. The base of the tower domain forms a direct connection to the oxidase domain and plays a crucial role in the shape and catalytic activity of the active site and removing the tower domain via a mutation resulted in a decrease in catalytic efficiency. <ref name="Stavropolous"/> The <scene name='83/834203/Phe538-tyr761interaction/5'>TαB-αD interaction</scene> is responsible for positioning of Phe538 in the catalytic chamber, where it is proposed to orient the substrate lysine through hydrophobic interaction. In addition, the TαB-αD interaction positions αD in the correct manner to provide a hydrogen bonding to Tyr761. Tyr761 is positioned in the catalytic chamber next to the FAD cofactor and aids in the binding of the lysine substrate. <ref name="Stavropolous"/> The tower domain has also been found to interact with other proteins and complexes, such as CoREST (Figure 2), as a switch to allosterically regulate the catalytic activity of the protein.<ref name="Yang">doi: 10.1016/j.molcel.2006.07.012</ref> | + | The <scene name='83/834203/Towerdomain/3'>tower domain</scene> is a 100 residue protrusion off of the main protein body that is caused by an insertion into the sequence of the oxidase domain. It is comprised of a coiled-coil of two [https://en.wikipedia.org/wiki/Alpha_helix α-helices]. The longer helix, TαA, is an LSD-1 specific element that has not been found in other oxidase proteins.<ref name="Stavropolous"/> The shorter helix, TαB, connects directly to helix αD of the oxidase domain through a conserved connector loop. The base of the tower domain forms a direct connection to the oxidase domain and plays a crucial role in the shape and catalytic activity of the active site and removing the tower domain via a mutation resulted in a decrease in catalytic efficiency.<ref name="Stavropolous"/> The <scene name='83/834203/Phe538-tyr761interaction/5'>TαB-αD interaction</scene> is responsible for positioning of Phe538 in the catalytic chamber, where it is proposed to orient the substrate lysine through hydrophobic interaction. In addition, the TαB-αD interaction positions αD in the correct manner to provide a hydrogen bonding to Tyr761. Tyr761 is positioned in the catalytic chamber next to the FAD cofactor and aids in the binding of the lysine substrate.<ref name="Stavropolous"/> The tower domain has also been found to interact with other proteins and complexes, such as CoREST (Figure 2), as a switch to allosterically regulate the catalytic activity of the protein.<ref name="Yang">doi: 10.1016/j.molcel.2006.07.012</ref> |
=== SWIRM Domain === | === SWIRM Domain === | ||
Revision as of 13:03, 3 August 2020
Human lysine-specific demethylase 1 (LSD-1), A repressor of transcription
| |||||||||||
Student Contributors
Nicholas Bantz, Sean Callahan, Cody Carley, Andrew Hesterhagen, Steve Klimcak, Michael Thomas
Proteopedia Page Contributors and Editors (what is this?)
Mark Macbeth, Valentine J Klimkowski, Michal Harel, Angel Herraez