Histone acetyltransferase 1-2 Complex (HAT1/2)
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
The structure of the <scene name='83/834210/Overview/2'>HAT1-HAT2 complex</scene> from the yeast ''Saccharomyces cerevisiae'' was determined by X-ray diffraction at 2.0Å resolution. The complex was crystallized in the presence of coenzyme A ([https://en.wikipedia.org/wiki/Coenzyme_A CoA]) and the histone H4 N-terminal peptide (residues 1-48). The complex has four components (HAT1, HAT2, H4 peptide, and CoA) seen with a 1:1:1:1 stoichiometry. While residues 1-48 of the yeast H4 protein were included in the crystallization, only residues 9-46 were well resolved in the refined structure. Similarly, residues 1-4 in the HAT1 subunit and residues 1-6, 87-104, and 391-401 in the HAT2 subunit were not resolved.<ref name="Yang"/> | The structure of the <scene name='83/834210/Overview/2'>HAT1-HAT2 complex</scene> from the yeast ''Saccharomyces cerevisiae'' was determined by X-ray diffraction at 2.0Å resolution. The complex was crystallized in the presence of coenzyme A ([https://en.wikipedia.org/wiki/Coenzyme_A CoA]) and the histone H4 N-terminal peptide (residues 1-48). The complex has four components (HAT1, HAT2, H4 peptide, and CoA) seen with a 1:1:1:1 stoichiometry. While residues 1-48 of the yeast H4 protein were included in the crystallization, only residues 9-46 were well resolved in the refined structure. Similarly, residues 1-4 in the HAT1 subunit and residues 1-6, 87-104, and 391-401 in the HAT2 subunit were not resolved.<ref name="Yang"/> | ||
| - | HAT1 is not catalytically active until it binds with HAT2 to form the complex.<ref name="Wu"> PMID: 22615379 </ref> The <scene name='83/834210/Hat1_-_chain_a/2'>HAT1 structure</scene> includes 317 residues and contains the binding site for acetyl | + | HAT1 is not catalytically active until it binds with HAT2 to form the complex.<ref name="Wu"> PMID: 22615379 </ref> The <scene name='83/834210/Hat1_-_chain_a/2'>HAT1 structure</scene> includes 317 residues and contains the binding site for acetyl coenzyme A. <scene name='83/834210/Hat2_-_chain_b/2'>HAT2</scene> is 401 residues and forms a beta-propeller structure with C7 symmetry. Bound to the HAT1-HAT2 complex is a <scene name='83/834210/Histone_4/3'>peptide substrate</scene> comprised of residues 9-46 of the histone protein H4. |
The HAT1 and HAT2 <scene name='81/811717/Salt_bridges/4'>interface</scene> is stabilized by multiple salt bridges, hydrogen bonds and van der Waals forces among hydrophobic side chains. Many of these are between a short helix spanning <scene name='83/834210/Lp1/3'>residues 200-208</scene> in HAT1 and several loops connecting β-strands in the propeller of HAT2. The HAT1 helix is important for the heterodimer formation as its deletion abolishes the ''in vitro'' interaction between HAT1 and HAT2.<ref name="Yang" /> Outside of the helix, three specific interactions at the interface involving hydrogen bonds help stabilize the complex: 1) The side chain atoms of <scene name='83/834210/Tyr199_asp308_ala202/4'>Tyr199 and Asp308</scene> with the main chain nitrogen of Ala202 in HAT1, 2) The side chain of <scene name='83/834210/Lys211phe205_and_leu288arg282/3'>Lys211 and Arg282</scene> with main chain atoms of Leu288 and Phe205 respectively, and 3) Between the side-chains of <scene name='83/834210/Serine_hydrogen_bonds/2'>Ser263 and Asp 206</scene>. Finally, the <scene name='83/834210/Hydrophobic_core/2'>hydrophobic core</scene> at the interface of the complex appears to be critical for the complex formation. This core consists of aromatic residues from HAT1 and leucine residues from HAT2, however it does not form any obvious ring stacking. | The HAT1 and HAT2 <scene name='81/811717/Salt_bridges/4'>interface</scene> is stabilized by multiple salt bridges, hydrogen bonds and van der Waals forces among hydrophobic side chains. Many of these are between a short helix spanning <scene name='83/834210/Lp1/3'>residues 200-208</scene> in HAT1 and several loops connecting β-strands in the propeller of HAT2. The HAT1 helix is important for the heterodimer formation as its deletion abolishes the ''in vitro'' interaction between HAT1 and HAT2.<ref name="Yang" /> Outside of the helix, three specific interactions at the interface involving hydrogen bonds help stabilize the complex: 1) The side chain atoms of <scene name='83/834210/Tyr199_asp308_ala202/4'>Tyr199 and Asp308</scene> with the main chain nitrogen of Ala202 in HAT1, 2) The side chain of <scene name='83/834210/Lys211phe205_and_leu288arg282/3'>Lys211 and Arg282</scene> with main chain atoms of Leu288 and Phe205 respectively, and 3) Between the side-chains of <scene name='83/834210/Serine_hydrogen_bonds/2'>Ser263 and Asp 206</scene>. Finally, the <scene name='83/834210/Hydrophobic_core/2'>hydrophobic core</scene> at the interface of the complex appears to be critical for the complex formation. This core consists of aromatic residues from HAT1 and leucine residues from HAT2, however it does not form any obvious ring stacking. | ||
| Line 23: | Line 23: | ||
===Acetyl-CoA Binding Site=== | ===Acetyl-CoA Binding Site=== | ||
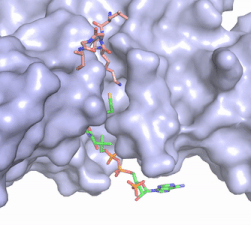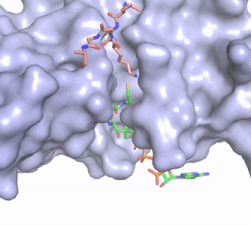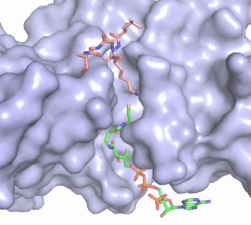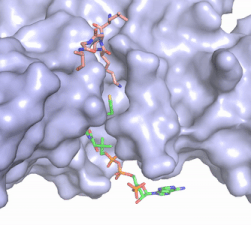
| - | [[Image:Hat1.gif|400 px|left|thumb|Figure 2. Coenzyme A (CoA, green sticks) and the histone H4 peptide substrate (pink sticks) are shown in the binding groove of HAT1. The movie was made in PyMol using 4psw.pdb, then converted to gif format using EZgif.]] The CoA molecule is buried in a deep channel in the HAT1 subunit, where the free rotation of several bonds in the [https://en.wikipedia.org/wiki/Pantetheine pantetheine] group give the molecule a bent conformation (Figure 2). The conserved Arg/Gln-X-X-Gly-X-Gly/Ala <scene name='83/834210/Newcoa/11'>signature sequence</scene> (residues 227-232) of CoA containing proteins wraps around the 5'-diphosphate moiety of the CoA. This motif also positions the negatively charged β-phosphate group of CoA at the N-terminal dipole of the helix spanning residues 230-245, further stabilizing the interaction. The <scene name='83/834210/Hydrophobic_pocket/2'>β-mercaptoethylamine group</scene> of CoA rests in the hydrophobic pocket formed by the side chains of residues Ile-217,Pro-257 and Phe-261. In the HAT1- | + | [[Image:Hat1.gif|400 px|left|thumb|Figure 2. Coenzyme A (CoA, green sticks) and the histone H4 peptide substrate (pink sticks) are shown in the binding groove of HAT1. The movie was made in PyMol using 4psw.pdb, then converted to gif format using EZgif.]] The CoA molecule is buried in a deep channel in the HAT1 subunit, where the free rotation of several bonds in the [https://en.wikipedia.org/wiki/Pantetheine pantetheine] group give the molecule a bent conformation (Figure 2). The conserved Arg/Gln-X-X-Gly-X-Gly/Ala <scene name='83/834210/Newcoa/11'>signature sequence</scene> (residues 227-232) of CoA containing proteins wraps around the 5'-diphosphate moiety of the CoA. This motif also positions the negatively charged β-phosphate group of CoA at the N-terminal dipole of the helix spanning residues 230-245, further stabilizing the interaction. The <scene name='83/834210/Hydrophobic_pocket/2'>β-mercaptoethylamine group</scene> of CoA rests in the hydrophobic pocket formed by the side chains of residues Ile-217,Pro-257 and Phe-261. In the HAT1-Acetyl coenzyme A structure determined without HAT2 and the H4 substrate (PDB: 1bob) the main chain atoms of <scene name='83/834210/Acetylcoa_structure/3'>Phe220</scene> were revealed to play a critical role in binding to acetyl-CoA. The carbonyl oxygen of Phe220 hydrogen bonds to the amine of the β-mercaptoethylamine gruop of the co-factor, while the Phe220 amide nitrogen hydrogen bonds to the acetyl group of acetyl-CoA.<ref name=”Dutnall”>PMID:10384314</ref> The latter interaction has important implications for the mechanism as described below. |
= Mechanism = | = Mechanism = | ||
| Line 29: | Line 29: | ||
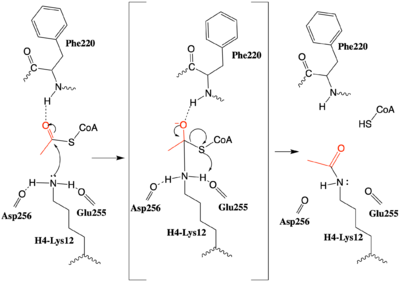
After many structural studies, the complete catalytic mechanism for HAT1 remains unclear. In particular, the identity of the general base needed to deprotonate the substrate lysine is uncertain. In a previous study a structural overlay of HAT1 and Gcn5, a better-understood HAT enzyme, found a conserved glutamate residue in the active site of both enzymes. Mutation of this glutamate (equivalent to Glu255 in 2psw.pdb) was shown to decrease the catalytic ability of HAT1, identifying it to be important for catalysis. <ref name="Yang"/> Given the proximity of <scene name='83/834210/Mechanism_glu_lys_coa/4'>Glu255 and Asp256</scene> this mechanism could be supported by structure of the HAT1-HAT2 complex with histone H4, however it would require a 180° shift in the direction of the side chains to act as a general base that deprotonates H4-Lys12, enhancing its nucleophilic character. | After many structural studies, the complete catalytic mechanism for HAT1 remains unclear. In particular, the identity of the general base needed to deprotonate the substrate lysine is uncertain. In a previous study a structural overlay of HAT1 and Gcn5, a better-understood HAT enzyme, found a conserved glutamate residue in the active site of both enzymes. Mutation of this glutamate (equivalent to Glu255 in 2psw.pdb) was shown to decrease the catalytic ability of HAT1, identifying it to be important for catalysis. <ref name="Yang"/> Given the proximity of <scene name='83/834210/Mechanism_glu_lys_coa/4'>Glu255 and Asp256</scene> this mechanism could be supported by structure of the HAT1-HAT2 complex with histone H4, however it would require a 180° shift in the direction of the side chains to act as a general base that deprotonates H4-Lys12, enhancing its nucleophilic character. | ||
| - | An alternative mechanism (Figure 3) that is better supported by the HAT1-HAT2-histone H4 structure, proposes that the attacking lysine would be deprotonated upon entry into active site due the proximity of the acidic side chains of Glu255 and Asp256. Additionally, there are <scene name='83/834210/Mechanism_lys_carbonyls/3'>three carbonyl oxygens</scene> in the main chain of Ser218, Glu255, and Asp256 which are in hydrogen bonding distance with the ε-amine of the attacking lysine. They will act to withdraw positive charge from the attacking nitrogen improving its nucleophilic nature and perhaps better orient the lone pair electrons for nucleophillic attack. Additionally, the interaction of the Phe220 amide with the carbonyl oxygen of the acetyl group enhances the electrophilic nature the carbonyl carbon being attacked. In the second step of the reaction, the lone pair on the lysine attacks the carbonyl carbon of acetyl-CoA, forming an oxyanion containing tetrahedral transition state. The structure does not definitively reveal residues in an oxyanion hole that stabilize the transition state [[Image:HAT1_mechanism.png|400px|right|thumb|Figure 3: The proposed HAT1 Mechanism with the transferred acetyl group in red. (The carbonyl of Ser218 described in the text and <scene name='83/834210/Mechanism_lys_carbonyls/1'>green link</scene> is not shown activating the Lys12 nucleophile)]] but superposition of the HAT1- | + | An alternative mechanism (Figure 3) that is better supported by the HAT1-HAT2-histone H4 structure, proposes that the attacking lysine would be deprotonated upon entry into active site due the proximity of the acidic side chains of Glu255 and Asp256. Additionally, there are <scene name='83/834210/Mechanism_lys_carbonyls/3'>three carbonyl oxygens</scene> in the main chain of Ser218, Glu255, and Asp256 which are in hydrogen bonding distance with the ε-amine of the attacking lysine. They will act to withdraw positive charge from the attacking nitrogen improving its nucleophilic nature and perhaps better orient the lone pair electrons for nucleophillic attack. Additionally, the interaction of the Phe220 amide with the carbonyl oxygen of the acetyl group enhances the electrophilic nature the carbonyl carbon being attacked. In the second step of the reaction, the lone pair on the lysine attacks the carbonyl carbon of acetyl-CoA, forming an oxyanion containing tetrahedral transition state. The structure does not definitively reveal residues in an oxyanion hole that stabilize the transition state [[Image:HAT1_mechanism.png|400px|right|thumb|Figure 3: The proposed HAT1 Mechanism with the transferred acetyl group in red. (The carbonyl of Ser218 described in the text and <scene name='83/834210/Mechanism_lys_carbonyls/1'>green link</scene> is not shown activating the Lys12 nucleophile)]] but superposition of the HAT1-acetyl coenzyme A structure with the HAT1-HAT2-H4 substrate structure suggests <scene name='83/834210/Mechanism_final/5'>the main chain amide of Phe220</scene>, which binds the carbonyl oxygen of the acetyl group before attack, is a likely candidate. Finally, upon electron reorganization, the C-S scissile bond breaks leaving the H4Lys12 acetylated and Coenzyme A as products. |
= Inhibition = | = Inhibition = | ||
Revision as of 16:07, 14 August 2020
The Yeast HAT1-HAT2 Histone Acetyltransferase Complex Bound to the Histone H4 substrate
| |||||||||||
Student Contributors
Morgan Buckley, Jordan Finch, Caitlin Gaich, Kiran Kaur, Emily Leiderman, Ben Nick
Proteopedia Page Contributors and Editors (what is this?)
Mark Macbeth, Angel Herraez, Michal Harel, Valentine J Klimkowski