We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Jeremiah C Hagler/Protein Visualization Lab COVID
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
The most important rule about protein structure is that it is determined by the primary sequence of the protein. Protein folding is a complicated multi-step process. The first step results in the '''secondary structure''' (or 2o structure) of the protein. Secondary structures come in two flavors: '''alpha helices''' and '''beta sheets''' (or beta-pleated sheets). Alpha helices are spiral staircase structures (see structure 1 below), and beta-pleated sheets are flat regions where the amino acids run back and forth next to each other in long ribbons (see structure 2 below). These two structures form spontaneously based on the shape/'''hydrophobicity'''/'''charges''' of the amino acids and are held together by '''hydrogen bonds'''. The protein will now look like a string of pearls with twists or zig-zags at intervals along its length. | The most important rule about protein structure is that it is determined by the primary sequence of the protein. Protein folding is a complicated multi-step process. The first step results in the '''secondary structure''' (or 2o structure) of the protein. Secondary structures come in two flavors: '''alpha helices''' and '''beta sheets''' (or beta-pleated sheets). Alpha helices are spiral staircase structures (see structure 1 below), and beta-pleated sheets are flat regions where the amino acids run back and forth next to each other in long ribbons (see structure 2 below). These two structures form spontaneously based on the shape/'''hydrophobicity'''/'''charges''' of the amino acids and are held together by '''hydrogen bonds'''. The protein will now look like a string of pearls with twists or zig-zags at intervals along its length. | ||
| - | |||
1. '''Three views of an alpha helix''' (In the structure window, click on "Popup" button to open a larger popup window of this structure. You can toggle the spin of the structure on or off by clicking on the "Spin" button. Clicking and holding on the structure in the window will allow you to manipulate the structure, rotating in three-dimensions.): | 1. '''Three views of an alpha helix''' (In the structure window, click on "Popup" button to open a larger popup window of this structure. You can toggle the spin of the structure on or off by clicking on the "Spin" button. Clicking and holding on the structure in the window will allow you to manipulate the structure, rotating in three-dimensions.): | ||
<br> | <br> | ||
a. <scene name='71/713432/Protein_secondary_structure/3'>Click to see alpha helix</scene> | a. <scene name='71/713432/Protein_secondary_structure/3'>Click to see alpha helix</scene> | ||
<br> | <br> | ||
| - | This is an alpha helix shown in isolation. The '''amino acid backbone''' (the parts of the amino acids that are linked together by a '''peptide bond''' to form the 1° sequence) is shown in pink/red. The '''amino acid side chains''' are shown in yellow (each type of amino acid has its own unique side chain, one of 20 different types). If a section of a protein's primary sequence of amino acids forms this coiled structure, it is known as an alpha-helix. | + | This is an '''alpha helix''' shown in isolation. The '''amino acid backbone''' (the parts of the amino acids that are linked together by a '''peptide bond''' to form the 1° sequence) is shown in pink/red. The '''amino acid side chains''' are shown in yellow (each type of amino acid has its own unique side chain, one of 20 different types). If a section of a protein's primary sequence of amino acids forms this coiled structure, it is known as an alpha-helix. |
| + | <br> | ||
b. <scene name='79/795987/Pg/9'>Click to see alpha helix in relation to beta sheet</scene> | b. <scene name='79/795987/Pg/9'>Click to see alpha helix in relation to beta sheet</scene> | ||
<br> | <br> | ||
| Line 23: | Line 23: | ||
c. <scene name='79/795987/Pg/5'>Click to see alpha helix highlighted</scene> | c. <scene name='79/795987/Pg/5'>Click to see alpha helix highlighted</scene> | ||
<br> | <br> | ||
| - | Here the alpha helix is red/pink, while adjoining amino acids from the primary sequence are shown in white. Again, the nearby beta-pleated sheet is shown as gold ribbon. | + | Here the alpha helix is red/pink, while adjoining amino acids from the primary sequence are shown in white. Again, the nearby beta-pleated sheet is shown as gold ribbon. |
| - | + | <br> | |
| - | 2. | + | <br> |
| + | 2. '''The Beta-pleated sheet''' | ||
<br> | <br> | ||
a. <scene name='71/713432/Protein_secondary_structure_bs/2'>Click to see beta sheet</scene> | a. <scene name='71/713432/Protein_secondary_structure_bs/2'>Click to see beta sheet</scene> | ||
<br> | <br> | ||
| - | Here the four parallel strings of amino acids are arrayed in a beta-pleated sheet. The protein backbone is red/pink, and side chains are shown in yellow. Each individual strand of the sheet is held in place by hydrogen bond interactions with the parallel strands of the sheet. | + | Here the four parallel strings of amino acids are arrayed in a '''beta-pleated sheet'''. The protein backbone is red/pink, and side chains are shown in yellow. Each individual strand of the sheet is held in place by hydrogen bond interactions with the parallel strands of the sheet. |
| + | <br> | ||
<br> | <br> | ||
The second step of protein folding results in the '''tertiary structure''' (or 3° structure). Tertiary structure gives the protein an overall three-dimensional structure. The tertiary structure of a protein is determined by a combination of factors including hydrogen bonds, '''ionic bonds''' (between positively and negatively charged amino acids), '''covalent''' '''disulfide bonds''' (between cysteine residues), and '''Van der Waals''' interactions. Tertiary structure can also be affected by repulsive forces between similarly charged amino acids, as well as '''hydrophobic''' and '''hydrophilic''' interactions with a solvent (commonly water). At a distance many proteins form what look to be large globs at this point, and it is only upon more careful and close up inspection that one can see the true uniqueness of the shape. | The second step of protein folding results in the '''tertiary structure''' (or 3° structure). Tertiary structure gives the protein an overall three-dimensional structure. The tertiary structure of a protein is determined by a combination of factors including hydrogen bonds, '''ionic bonds''' (between positively and negatively charged amino acids), '''covalent''' '''disulfide bonds''' (between cysteine residues), and '''Van der Waals''' interactions. Tertiary structure can also be affected by repulsive forces between similarly charged amino acids, as well as '''hydrophobic''' and '''hydrophilic''' interactions with a solvent (commonly water). At a distance many proteins form what look to be large globs at this point, and it is only upon more careful and close up inspection that one can see the true uniqueness of the shape. | ||
| Line 38: | Line 40: | ||
Proteins may contain only alpha helices, only beta sheets, or a combination of the two. The same holds true for the bonds giving a protein its tertiary structure - all, some or none may be present. These different folding patterns existing in different proteins are what give the proteins their distinctive shapes and sizes. A protein that is 300 amino acids long will be 100 nm as an extended chain. If the protein is an alpha helix, it will be 45 nm long; a beta sheet will be 7 x 7 x 0.8 nm; and a small globular form will form a sphere only 4.5 nm in diameter! | Proteins may contain only alpha helices, only beta sheets, or a combination of the two. The same holds true for the bonds giving a protein its tertiary structure - all, some or none may be present. These different folding patterns existing in different proteins are what give the proteins their distinctive shapes and sizes. A protein that is 300 amino acids long will be 100 nm as an extended chain. If the protein is an alpha helix, it will be 45 nm long; a beta sheet will be 7 x 7 x 0.8 nm; and a small globular form will form a sphere only 4.5 nm in diameter! | ||
| - | Finally, multiple proteins might interact to form a quaternary structure (4°), sometimes known as a protein complex. Each protein in a quaternary structure is called a subunit, and in some cases multiple subunits of the same protein interact to form a quaternary structure (as in the SARS-CoV-2 spike protein you will learn more about below). On the other hands, some quaternary structures contain multiple types of proteins that interact. Most quaternary structures are held together by weak intermolecular bonds (ionic, H-bonds, hydrophobic, van der waals, etc) and occasionally by strong covalent bonds (such as disulfide bonds). While the designation "subunit" is usually reserved for a individual protein that makes up a part of a larger quaternary structure, in some cases term subunit is | + | Finally, multiple proteins might interact to form a quaternary structure (4°), sometimes known as a protein complex. Each protein in a quaternary structure is called a subunit, and in some cases multiple subunits of the same protein interact to form a quaternary structure (as in the SARS-CoV-2 spike protein you will learn more about below). On the other hands, some quaternary structures contain multiple types of proteins that interact. Most quaternary structures are held together by weak intermolecular bonds (ionic, H-bonds, hydrophobic, van der waals, etc) and occasionally by strong covalent bonds (such as disulfide bonds). While the designation "subunit" is usually reserved for a individual protein that makes up a part of a larger quaternary structure, in some cases the term subunit is also be used to designate large portions of a protein structure that have distinct functions. An example of this alternative use is found in the SARS-CoV-2 spike protein (below). Even though each spike protein is made up of three identical subunits (thus is a homotrimer), each of these single polypeptide is further divided into two main subunits--the receptor-binding subunit (S1) and membrane-fusion subunit (S2) (see below). |
Domains: | Domains: | ||
Parts of the secondary and tertiary structures of a protein are usually arranged to form domains, functional units associated with a particular structure. For example, a pair of alpha helices situated side by side might form a binding site, or a particular folding pattern might form the active site of an enzyme, where it binds to its substrate, or the site at which it binds to a coenzyme such as NAD+. The structure of the domain (though not necessarily the exact amino acid sequence) is frequently preserved in different proteins from the same organism that have a similar function (to bind to a cellular receptor, for example, in the SARS-CoV-2 Spike Protein, or to move phosphate groups, for instance). Domains are also conserved in proteins from different species that have the same function (such as hemoglobins for oxygen transport or cytochromes in the electron transfer system of mitochondria). Variations in the amino acid sequences in similar domains (or in the nucleotide sequences or genes that code for the proteins) give important clues about evolutionary relationships between organisms. Individual domains are sometimes found (but not always, a fact that makes this a very controversial topic) contained within single exons of eukaryotic genes (exons and introns are concepts you will learn more about later in the course, when we discuss eukaryotic gene structure). In other words, a single exon might represent all of the protein coding sequence required to generate a functional domain within the context of the whole protein structure. This finding has implications for the evolution of eukaryotic genes, since it implies that new proteins can be generated by simply duplicating preexisting protein domain encoding exons and recombining them into new combinations (a process known as exon-shuffling). Thus, a vast variety of proteins with new functions can be generated from preexisting genes, allowing great evolutionary flexibility. Looking at the genes of many eukaryotic organisms shows that this is exactly what appears to happen. | Parts of the secondary and tertiary structures of a protein are usually arranged to form domains, functional units associated with a particular structure. For example, a pair of alpha helices situated side by side might form a binding site, or a particular folding pattern might form the active site of an enzyme, where it binds to its substrate, or the site at which it binds to a coenzyme such as NAD+. The structure of the domain (though not necessarily the exact amino acid sequence) is frequently preserved in different proteins from the same organism that have a similar function (to bind to a cellular receptor, for example, in the SARS-CoV-2 Spike Protein, or to move phosphate groups, for instance). Domains are also conserved in proteins from different species that have the same function (such as hemoglobins for oxygen transport or cytochromes in the electron transfer system of mitochondria). Variations in the amino acid sequences in similar domains (or in the nucleotide sequences or genes that code for the proteins) give important clues about evolutionary relationships between organisms. Individual domains are sometimes found (but not always, a fact that makes this a very controversial topic) contained within single exons of eukaryotic genes (exons and introns are concepts you will learn more about later in the course, when we discuss eukaryotic gene structure). In other words, a single exon might represent all of the protein coding sequence required to generate a functional domain within the context of the whole protein structure. This finding has implications for the evolution of eukaryotic genes, since it implies that new proteins can be generated by simply duplicating preexisting protein domain encoding exons and recombining them into new combinations (a process known as exon-shuffling). Thus, a vast variety of proteins with new functions can be generated from preexisting genes, allowing great evolutionary flexibility. Looking at the genes of many eukaryotic organisms shows that this is exactly what appears to happen. | ||
| + | <br> | ||
A good example of all of these principles can be found in the SARS-CoV-2 Spike protein (see figure 4). This is the protein the SARS-CoV-2 virus relies on to gain entrance into its target......that target being any cell that contains the Angiotensin-converting enzyme 2 (ACE2) protein on its surface. The Spike protein is made up of three identical strands of protein arranged into a quaternary structure (a homotrimer). Each is synthesized as an individual protein and then later complexed into the complex secondary, tertiary and quaternary structure you see below. Each individual polypeptide of the spike protein is divided into four major components, S1 (or receptor-binding subunit), S2 (membrane-fusion subunit), TM (trans-membrane anchor) and IC (intracellular-tail). The interaction of secondary structure to form tertiary structure, and the interaction of these structures to form quaternary structure is apparent. The final step of protein folding results in quarternary structure (or 4° structure). This step is only taken in proteins that are made of multiple subunits; meaning that strands of proteins - coded for on separate mRNAs and synthesized independently - come together to form a single functional molecule. Many proteins have multiple subunits; for example, the SAR-CoV-2 protein is made up of three subunits (figure 4). | A good example of all of these principles can be found in the SARS-CoV-2 Spike protein (see figure 4). This is the protein the SARS-CoV-2 virus relies on to gain entrance into its target......that target being any cell that contains the Angiotensin-converting enzyme 2 (ACE2) protein on its surface. The Spike protein is made up of three identical strands of protein arranged into a quaternary structure (a homotrimer). Each is synthesized as an individual protein and then later complexed into the complex secondary, tertiary and quaternary structure you see below. Each individual polypeptide of the spike protein is divided into four major components, S1 (or receptor-binding subunit), S2 (membrane-fusion subunit), TM (trans-membrane anchor) and IC (intracellular-tail). The interaction of secondary structure to form tertiary structure, and the interaction of these structures to form quaternary structure is apparent. The final step of protein folding results in quarternary structure (or 4° structure). This step is only taken in proteins that are made of multiple subunits; meaning that strands of proteins - coded for on separate mRNAs and synthesized independently - come together to form a single functional molecule. Many proteins have multiple subunits; for example, the SAR-CoV-2 protein is made up of three subunits (figure 4). | ||
| Line 49: | Line 52: | ||
a. [[Image:PPC_Motif_in_SARS-CoV-2_Spike_Protein_Cropped_to_Spike_Schematic_Only_B.jpg|300px|]] | a. [[Image:PPC_Motif_in_SARS-CoV-2_Spike_Protein_Cropped_to_Spike_Schematic_Only_B.jpg|300px|]] | ||
<br> | <br> | ||
| - | Schematic drawing of the three-dimensional (3D) structure of SARS-CoV-2 coronavirus spike. S1, receptor-binding subunit; S2, membrane fusion subunit; TM, transmembrane anchor; IC, intracellular tail. | + | Schematic drawing of the three-dimensional (3D) structure of SARS-CoV-2 coronavirus spike. S1, receptor-binding subunit; S2, membrane fusion subunit; TM, transmembrane anchor; IC, intracellular tail.[modified from Figure 1 of [Shang et al.,PNAS May 26, 2020 117 (21) 11727-11734; first published May 6, 2020 https://doi.org/10.1073/pnas.2003138117]] |
<br> | <br> | ||
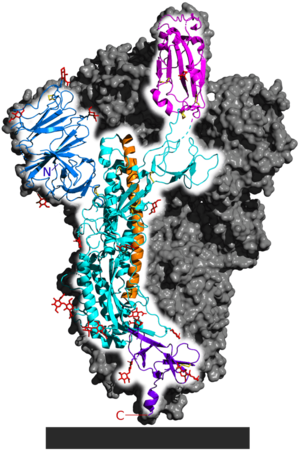
b. [[Image:716px-6VSB_spike_protein_SARS-CoV-2_monomer_in_homotrimer.png|6VSB spike protein SARS-CoV-2 monomer in homotrimer|300px]] | b. [[Image:716px-6VSB_spike_protein_SARS-CoV-2_monomer_in_homotrimer.png|6VSB spike protein SARS-CoV-2 monomer in homotrimer|300px]] | ||
<br> | <br> | ||
| - | This is the same structure represented in the schematic above, except the 3-dimensional structure of one of the three components of the overall spike protein quaternary structure is shown (the other two members of the homotrimer are grey) . The flat ribbons show beta-sheets while the cylindrical coils represent alpha-helices. The S1 receptor-binding subunit is magenta, while the S2 membrane-fusion subunit consists of the blue, teal, orange and purple portions. The TM and IC domains are not shown. | + | This is the same structure represented in the schematic above, except the 3-dimensional structure of one of the three components of the overall spike protein quaternary structure is shown (the other two members of the homotrimer are grey) . The flat ribbons show beta-sheets while the cylindrical coils represent alpha-helices. The S1 receptor-binding subunit is magenta, while the S2 membrane-fusion subunit consists of the blue, teal, orange and purple portions. The TM and IC domains are not shown.[from [https://commons.wikimedia.org/wiki/File:6VSB_spike_protein_SARS-CoV-2_monomer_in_homotrimer.png#file]] |
<br> | <br> | ||
<br> | <br> | ||
Revision as of 19:51, 24 September 2020
Introduction to Computer-Aided Protein Visualization Lab
| |||||||||||