Hypoxia-Inducible Factors
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | <StructureSection load='4ZPK' size=' | + | <StructureSection load='4ZPK' size='400' side='right' caption='Crystal structure of HIF-2α/HIF-1β Complex with HRE DNA (PDB code [https://www.rcsb.org/structure/4zpk 4ZPK])' > |
'''Hypoxia-inducible factors''' ('''HIFs''') are transcription factors responsible of the cellular adaptation to [https://en.wikipedia.org/wiki/Hypoxia_(medical) hypoxia], which is a condition of low oxygen availability. Among the genes regulated by HIF, we can find those involved in [https://en.wikipedia.org/wiki/Erythropoiesis erythropoiesis], [https://en.wikipedia.org/wiki/Angiogenesis angiogenesis] and [https://en.wikipedia.org/wiki/Metabolism metabolism]. | '''Hypoxia-inducible factors''' ('''HIFs''') are transcription factors responsible of the cellular adaptation to [https://en.wikipedia.org/wiki/Hypoxia_(medical) hypoxia], which is a condition of low oxygen availability. Among the genes regulated by HIF, we can find those involved in [https://en.wikipedia.org/wiki/Erythropoiesis erythropoiesis], [https://en.wikipedia.org/wiki/Angiogenesis angiogenesis] and [https://en.wikipedia.org/wiki/Metabolism metabolism]. | ||
| Line 35: | Line 35: | ||
==Physiological Function== | ==Physiological Function== | ||
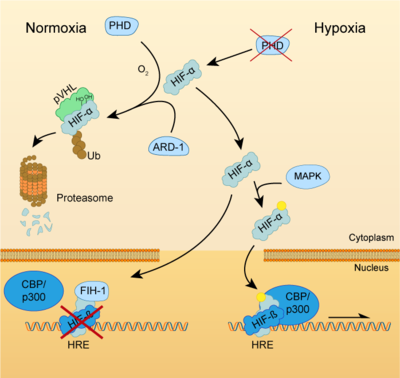
| + | [[Image:HIF-pathway-GonzaloGarcia-Martin-2020.png|400 px|thumb|right|HIF pathway]] | ||
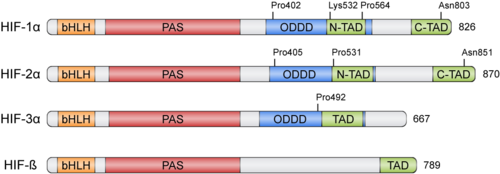
Both HIF-α and β subunits are constitutively and ubiquitously expressed in almost every single cell, although some isoforms may be predominant in some specific tissues<ref>PMID: 20396958</ref>. For example, HIF-2α is mostly expressed in the endothelium, kidney, lung, heart and small intestine . While HIF-β protein levels in the cell are constant, HIF-α has a short half-life (~ 5 mins) and is highly regulated by oxygen through its ODDD domain. With normal oxygen levels (normoxia), HIF-α protein levels are rapidly degraded, resulting in essentially no detectable HIF-α protein. However, in hypoxic conditions HIF-α becomes stabilized and is translocated from the cytoplasm to the nucleus, where it dimerizes with HIF-β and the HIF complex formed becomes transcriptionally active. More than 100 genes with varying functions have been described to be transcribed by the action of HIF. Moreover, HIF regulates up to 2% of all human genes in arterial endothelial cells, either directly or indirectly<ref>PMID: 15374877</ref>. In response to hypoxia, the capacity of red blood cells to transport oxygen is upregulated by the expression of genes like ''EPO'', essential in [https://en.wikipedia.org/wiki/Erythropoiesis erythropoiesis], and genes involved in iron metabolism. In addition, a large number of genes involved in different steps of [https://en.wikipedia.org/wiki/Angiogenesis angiogenesis] have been shown to increase by hypoxia challenge, being the [[Vascular Endothelial Growth Factor | '''Vascular Endothelial Growth Factor''']] ('''VEGF''') among them. VEGF directly recruits endothelial cells into hypoxic areas to generate new blood vessels by stimulating their proliferation. Furthermore, HIF complexes have been shown to induce pro-survival factors such as [[Insulin-like growth factor | '''Insulin-like Growth Factor-2''']] ('''IGF2''') and [https://en.wikipedia.org/wiki/TGF_alpha '''Transforming Growth Factor-α'''] ('''TGF-α'''), which in turn enhances expression of HIFα itself. | Both HIF-α and β subunits are constitutively and ubiquitously expressed in almost every single cell, although some isoforms may be predominant in some specific tissues<ref>PMID: 20396958</ref>. For example, HIF-2α is mostly expressed in the endothelium, kidney, lung, heart and small intestine . While HIF-β protein levels in the cell are constant, HIF-α has a short half-life (~ 5 mins) and is highly regulated by oxygen through its ODDD domain. With normal oxygen levels (normoxia), HIF-α protein levels are rapidly degraded, resulting in essentially no detectable HIF-α protein. However, in hypoxic conditions HIF-α becomes stabilized and is translocated from the cytoplasm to the nucleus, where it dimerizes with HIF-β and the HIF complex formed becomes transcriptionally active. More than 100 genes with varying functions have been described to be transcribed by the action of HIF. Moreover, HIF regulates up to 2% of all human genes in arterial endothelial cells, either directly or indirectly<ref>PMID: 15374877</ref>. In response to hypoxia, the capacity of red blood cells to transport oxygen is upregulated by the expression of genes like ''EPO'', essential in [https://en.wikipedia.org/wiki/Erythropoiesis erythropoiesis], and genes involved in iron metabolism. In addition, a large number of genes involved in different steps of [https://en.wikipedia.org/wiki/Angiogenesis angiogenesis] have been shown to increase by hypoxia challenge, being the [[Vascular Endothelial Growth Factor | '''Vascular Endothelial Growth Factor''']] ('''VEGF''') among them. VEGF directly recruits endothelial cells into hypoxic areas to generate new blood vessels by stimulating their proliferation. Furthermore, HIF complexes have been shown to induce pro-survival factors such as [[Insulin-like growth factor | '''Insulin-like Growth Factor-2''']] ('''IGF2''') and [https://en.wikipedia.org/wiki/TGF_alpha '''Transforming Growth Factor-α'''] ('''TGF-α'''), which in turn enhances expression of HIFα itself. | ||
Revision as of 10:16, 15 November 2020
| |||||||||||
References
- ↑ Ke Q, Costa M. Hypoxia-inducible factor-1 (HIF-1). Mol Pharmacol. 2006 Nov;70(5):1469-80. doi: 10.1124/mol.106.027029. Epub 2006 Aug, 3. PMID:16887934 doi:http://dx.doi.org/10.1124/mol.106.027029
- ↑ Wu D, Potluri N, Lu J, Kim Y, Rastinejad F. Structural integration in hypoxia-inducible factors. Nature. 2015 Aug 5. doi: 10.1038/nature14883. PMID:26245371 doi:http://dx.doi.org/10.1038/nature14883
- ↑ Goldberg MA, Dunning SP, Bunn HF. Regulation of the erythropoietin gene: evidence that the oxygen sensor is a heme protein. Science. 1988 Dec 9;242(4884):1412-5. doi: 10.1126/science.2849206. PMID:2849206 doi:http://dx.doi.org/10.1126/science.2849206
- ↑ Semenza GL, Nejfelt MK, Chi SM, Antonarakis SE. Hypoxia-inducible nuclear factors bind to an enhancer element located 3' to the human erythropoietin gene. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5680-4. doi: 10.1073/pnas.88.13.5680. PMID:2062846 doi:http://dx.doi.org/10.1073/pnas.88.13.5680
- ↑ Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995 Jun 6;92(12):5510-4. PMID:7539918
- ↑ Reyes H, Reisz-Porszasz S, Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science. 1992 May 22;256(5060):1193-5. PMID:1317062
- ↑ Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci U S A. 2002 Apr 16;99(8):5271-6. PMID:11959977 doi:http://dx.doi.org/10.1073/pnas.082121399
- ↑ Loboda A, Jozkowicz A, Dulak J. HIF-1 and HIF-2 transcription factors--similar but not identical. Mol Cells. 2010 May;29(5):435-42. doi: 10.1007/s10059-010-0067-2. Epub 2010 Apr, 12. PMID:20396958 doi:http://dx.doi.org/10.1007/s10059-010-0067-2
- ↑ Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005 Jan 15;105(2):659-69. doi: 10.1182/blood-2004-07-2958. Epub 2004 Sep , 16. PMID:15374877 doi:http://dx.doi.org/10.1182/blood-2004-07-2958
- ↑ Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001 Nov 9;294(5545):1337-40. doi: 10.1126/science.1066373. Epub 2001, Oct 11. PMID:11598268 doi:http://dx.doi.org/10.1126/science.1066373
- ↑ Kamura T, Sato S, Iwai K, Czyzyk-Krzeska M, Conaway RC, Conaway JW. Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc Natl Acad Sci U S A. 2000 Sep 12;97(19):10430-5. PMID:10973499 doi:http://dx.doi.org/10.1073/pnas.190332597
- ↑ Jeong JW, Bae MK, Ahn MY, Kim SH, Sohn TK, Bae MH, Yoo MA, Song EJ, Lee KJ, Kim KW. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell. 2002 Nov 27;111(5):709-20. PMID:12464182
- ↑ Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002 Feb 1;295(5556):858-61. doi: 10.1126/science.1068592. PMID:11823643 doi:http://dx.doi.org/10.1126/science.1068592
- ↑ Richard DE, Berra E, Gothie E, Roux D, Pouyssegur J. p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1alpha (HIF-1alpha) and enhance the transcriptional activity of HIF-1. J Biol Chem. 1999 Nov 12;274(46):32631-7. doi: 10.1074/jbc.274.46.32631. PMID:10551817 doi:http://dx.doi.org/10.1074/jbc.274.46.32631
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
Proteopedia Page Contributors and Editors (what is this?)
Gonzalo Garcia-Martin, Michal Harel, Carmen Paredes Yubero, Rocío Moreno