Sandbox GGC1
From Proteopedia
| Line 5: | Line 5: | ||
== Function == | == Function == | ||
| - | [https://www.uniprot.org/uniprot/P84243 histone H3] replaces H3 in a range of nucleosomes in active genes | + | [https://www.uniprot.org/uniprot/P84243 histone H3] replaces H3 in a range of nucleosomes in active genes and it takes over the original H3 in non dividing cells. Nucleosomes wrap around and compact DNA into chromatin which limits DNA access to cellular machineries which need DNA as a template. Histones play an important role in regulation of transcription, DNA repair, DNA replication and also chromosomal stability. Access to DNA is regulated by post-translational modifications of histones which is called a histone code, and nucleosome remodeling. It also serves as a replacement histone that's imbedded chromatin regions by the HIRA chaperone, after the depletion of the H3.1 during transcription and DNA repair. |
== Disease == | == Disease == | ||
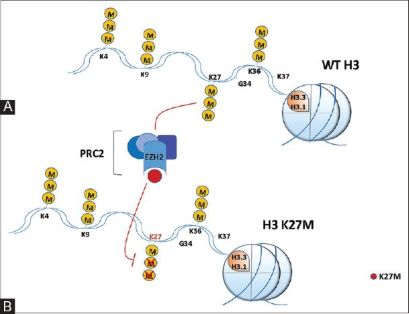
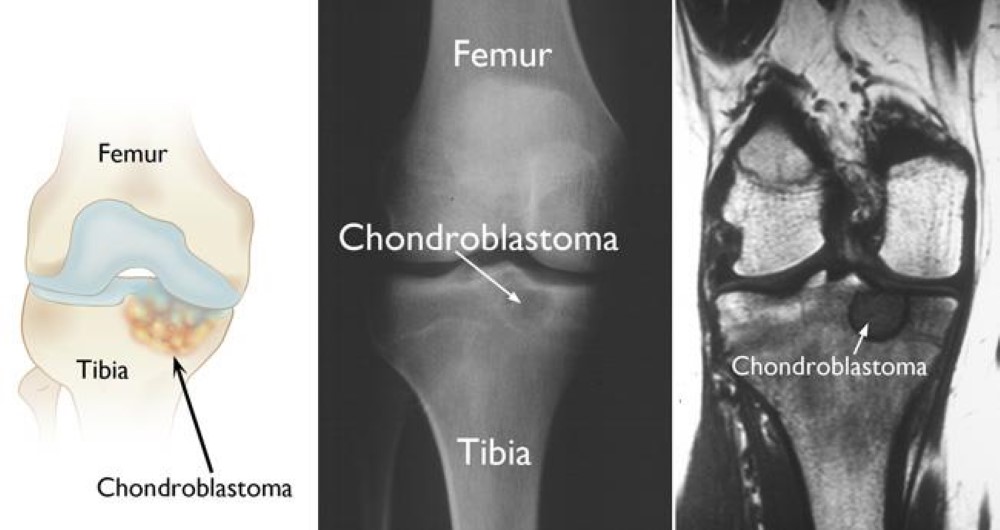
| - | There are mutations in H3.3 that are found in different types of bone tumors like [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4446520/ chrondroblastoma] for example and giant cell tumors of the bone. [https://cancerdiscovery.aacrjournals.org/content/3/12/1329.1 Chondroblastoma] arises in children and in young adults in the cartilage of the growth plates of the long bones and is most typically benign. | + | There have been studies that have identified mutations encoding a K27M substitution and there have also been mutations that encoded |
| + | GLY 34 to ARG or VAL called the G34R/V substitution. There are mutations in H3.3 that are found in different types of bone tumors like [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4446520/ chrondroblastoma] for example and giant cell tumors of the bone. [https://cancerdiscovery.aacrjournals.org/content/3/12/1329.1 Chondroblastoma] arises in children and in young adults in the cartilage of the growth plates of the long bones and is most typically benign. | ||
[[Image:Glioma_2018_1_4_117_240231_f1.jpg]] | [[Image:Glioma_2018_1_4_117_240231_f1.jpg]] | ||
| + | |||
| + | [[Image:a00609f01_resize.jpg]] | ||
== Relevance == | == Relevance == | ||
Revision as of 00:32, 16 November 2020
Histone H3.3
| |||||||||||
References
1. Arimura, Y.; Shirayama, K.; Horikoshi, N.; Fujita, R.; Taguchi, H.; Kagawa, W.; Fukagawa, T.; Almouzni, G.; Kurumizaka, H. Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3. https://www.nature.com/articles/srep07115 (accessed Nov 1, 2020).
2. Cancer Discovery Science Writers. Histone H3.3 Mutations Are Cancer Type-Specific. https://cancerdiscovery.aacrjournals.org/content/3/12/1329.1 (accessed Nov 14, 2020).
3. Kallappagoudar, S.; Yadav, R. K.; Lowe, B. R.; Partridge, J. F. Histone H3 mutations--a special role for H3.3 in tumorigenesis? https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4446520/ (accessed Nov 1, 2020).
4. UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. Histone H3.3. https://www.uniprot.org/uniprot/P84243 (accessed Nov 1, 2020).