We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox GGC3
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
==Domains== | ==Domains== | ||
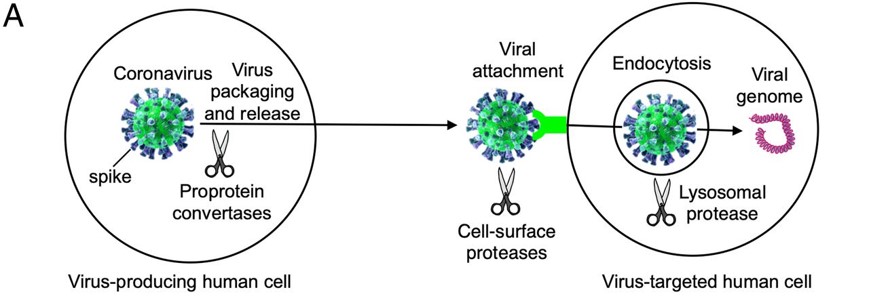
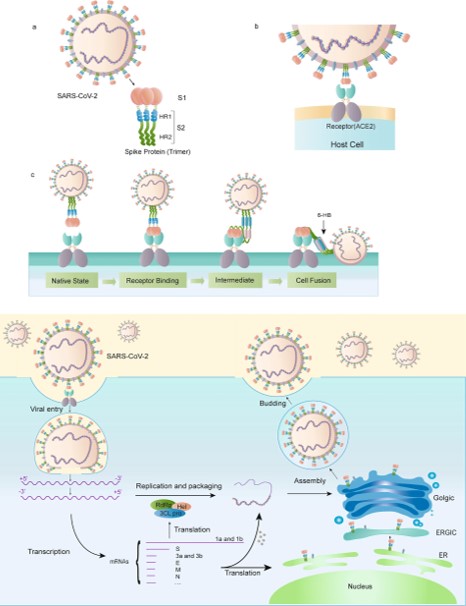
| - | The S-protein of the SARS-CoV-2 has two main subunits , an S1 and S2 subunits NTD-CT<scene name='75/752266/Ntd_-_ct/1'>S1 subunit is in the downstream of NTD and S2 subunit is in the upsteam of CT</scene> . The S1 subunit is located on residue #14–685 ( contains the NTD) and interact with human AEC2 by attaching its virion to the cell membrane by interacting with host receptor. The S2 subunit is located on residue #686–1273 ( contains the CT) which serves as the fusion protein of the virus <ref>UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. (2020, October 07). Spike glycoprotein. Retrieved November 13, 2020, from https://www.uniprot.org/uniprot/P59594</ref>. | + | The S-protein of the SARS-CoV-2 has two main subunits , an S1 and S2 subunits NTD-CT<scene name='75/752266/Ntd_-_ct/1'>S1 subunit is in the downstream of NTD and S2 subunit is in the upsteam of CT</scene> . The S1 subunit is located on residue #14–685 ( contains the NTD, blue label) and interact with human AEC2 by attaching its virion to the cell membrane by interacting with host receptor. The S2 subunit is located on residue #686–1273 ( contains the CT, red label ) which serves as the fusion protein of the virus <ref>UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. (2020, October 07). Spike glycoprotein. Retrieved November 13, 2020, from https://www.uniprot.org/uniprot/P59594</ref>. |
Revision as of 14:01, 16 November 2020
Spike glycoprotein
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Huang, Y., Yang, C., Xu, X., Xu, W., & Liu, S. (2020). Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacologica Sinica, 41(9), 1141-1149. doi:10.1038/s41401-020-0485-4
- ↑ Mohammad, A., Alshawaf, E., Marafie, S. K., Abu-Farha, M., Abubaker, J., & Al-Mulla, F. (2020). Higher binding affinity of Furin to SARS-CoV-2 spike (S) protein D614G could be associated with higher SARS-CoV-2 infectivity. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases, S1201-9712(20)32237-2. Advance online publication. https://doi.org/10.1016/j.ijid.2020.10.033
- ↑ UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. (2020, October 07). Spike glycoprotein. Retrieved November 13, 2020, from https://www.uniprot.org/uniprot/P59594
- ↑ Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., & Li, F. (2020, May 26). Cell entry mechanisms of SARS-CoV-2. Retrieved November 14, 2020, from https://www.pnas.org/content/117/21/11727
- ↑ Huang, Y., Yang, C., Xu, X., Xu, W., & Liu, S. (2020). Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacologica Sinica, 41(9), 1141-1149. doi:10.1038/s41401-020-0485-4
- ↑ Mohammad, A., Alshawaf, E., Marafie, S. K., Abu-Farha, M., Abubaker, J., & Al-Mulla, F. (2020). Higher binding affinity of Furin to SARS-CoV-2 spike (S) protein D614G could be associated with higher SARS-CoV-2 infectivity. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases, S1201-9712(20)32237-2. Advance online publication. https://doi.org/10.1016/j.ijid.2020.10.033