Sandbox GGC1
From Proteopedia
| Line 1: | Line 1: | ||
'''Histone H3.3'''<StructureSection load='3WTP' size='340' side='right' caption='Caption for this structure' scene=''> | '''Histone H3.3'''<StructureSection load='3WTP' size='340' side='right' caption='Caption for this structure' scene=''> | ||
== Introduction == | == Introduction == | ||
| - | Histone H3.3 is a variant histone of H3 which has the gene name H3.3A and this particular protein is found in Humans. the location of this can be found in the nucleus and in the chromosome. | + | Histone H3.3 is a variant histone of H3 which has the gene name H3.3A and this particular protein is found in Humans. the location of this can be found in the nucleus and in the chromosome.<ref>https://www.uniprot.org/uniprot/P84243</ref> |
== Function == | == Function == | ||
| - | [https://www.uniprot.org/uniprot/P84243 Histone H3] replaces H3 in a range of nucleosomes in active genes and it takes over the original H3 in non dividing cells. Nucleosomes wrap around and compact DNA into chromatin which limits DNA access to cellular machineries which need DNA as a template. Histones play an important role in regulation of transcription, DNA repair, DNA replication and also chromosomal stability. Access to DNA is regulated by post-translational modifications of histones which is called a histone code, and nucleosome remodeling. It also serves as a replacement histone that's imbedded chromatin regions by the HIRA chaperone, after the depletion of the H3.1 during transcription and DNA repair. | + | [https://www.uniprot.org/uniprot/P84243 Histone H3] replaces H3 in a range of nucleosomes in active genes and it takes over the original H3 in non dividing cells. Nucleosomes wrap around and compact DNA into chromatin which limits DNA access to cellular machineries which need DNA as a template. Histones play an important role in regulation of transcription, DNA repair, DNA replication and also chromosomal stability. Access to DNA is regulated by post-translational modifications of histones which is called a histone code, and nucleosome remodeling. It also serves as a replacement histone that's imbedded chromatin regions by the HIRA chaperone, after the depletion of the H3.1 during transcription and DNA repair.<ref>https://www.uniprot.org/uniprot/P84243</ref> |
== Disease == | == Disease == | ||
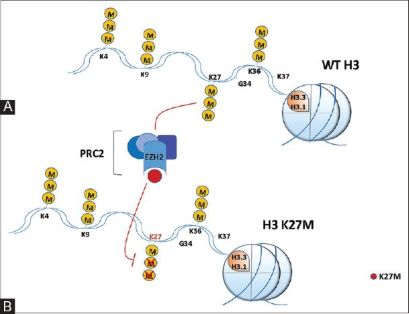
There have been studies that have identified mutations encoding a K27M substitution and there have also been mutations that encoded | There have been studies that have identified mutations encoding a K27M substitution and there have also been mutations that encoded | ||
| Line 15: | Line 15: | ||
== Relevance == | == Relevance == | ||
| + | |||
Histone octamer containing two of H2A, H2B, H3 and H4 and the octamer wraps 147bp of DNA. H3.3 interacts with HIRA which is a chaperone and ZMYND11 when trimethylated at <scene name='75/752263/3wtp/2'>Lysine-36</scene> (color coded dark blue). HIRA can deposit histones of H3.3 in replicating and non replicating cells. Also in the HIRA complex, Anti slicing factors ASF1a and ASF1b are purified by H3.3. However, when there's a disappearance of HIRA which is responsible for H3.3 deposition, it can cause defects in the early stages of embryogenesis. ZMYND11 also known as BS69 can find H3.3 LYS 36 (H3.3K36me3) by the binding domain of chromatin. | Histone octamer containing two of H2A, H2B, H3 and H4 and the octamer wraps 147bp of DNA. H3.3 interacts with HIRA which is a chaperone and ZMYND11 when trimethylated at <scene name='75/752263/3wtp/2'>Lysine-36</scene> (color coded dark blue). HIRA can deposit histones of H3.3 in replicating and non replicating cells. Also in the HIRA complex, Anti slicing factors ASF1a and ASF1b are purified by H3.3. However, when there's a disappearance of HIRA which is responsible for H3.3 deposition, it can cause defects in the early stages of embryogenesis. ZMYND11 also known as BS69 can find H3.3 LYS 36 (H3.3K36me3) by the binding domain of chromatin. | ||
== Structural highlights == | == Structural highlights == | ||
| Line 21: | Line 22: | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| + | <references /> | ||
1. Arimura, Y.; Shirayama, K.; Horikoshi, N.; Fujita, R.; Taguchi, H.; Kagawa, W.; Fukagawa, T.; Almouzni, G.; Kurumizaka, H. Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3. https://www.nature.com/articles/srep07115 (accessed Nov 1, 2020). | 1. Arimura, Y.; Shirayama, K.; Horikoshi, N.; Fujita, R.; Taguchi, H.; Kagawa, W.; Fukagawa, T.; Almouzni, G.; Kurumizaka, H. Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3. https://www.nature.com/articles/srep07115 (accessed Nov 1, 2020). | ||
Revision as of 15:18, 16 November 2020
Histone H3.3
| |||||||||||
References
1. Arimura, Y.; Shirayama, K.; Horikoshi, N.; Fujita, R.; Taguchi, H.; Kagawa, W.; Fukagawa, T.; Almouzni, G.; Kurumizaka, H. Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3. https://www.nature.com/articles/srep07115 (accessed Nov 1, 2020).
2. Cancer Discovery Science Writers. Histone H3.3 Mutations Are Cancer Type-Specific. https://cancerdiscovery.aacrjournals.org/content/3/12/1329.1 (accessed Nov 14, 2020).
3. Gianno, F.; Antonelli, M.; Ferretti2018, E.; Massimino, M.; Arcella, A.; Giangaspero, F. Pediatric high-grade glioma: A heterogeneous group of neoplasms with different molecular drivers.  (accessed Nov 16, 2020).
(accessed Nov 16, 2020).
4. Kallappagoudar, S.; Yadav, R. K.; Lowe, B. R.; Partridge, J. F. Histone H3 mutations--a special role for H3.3 in tumorigenesis? https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4446520/ (accessed Nov 1, 2020).
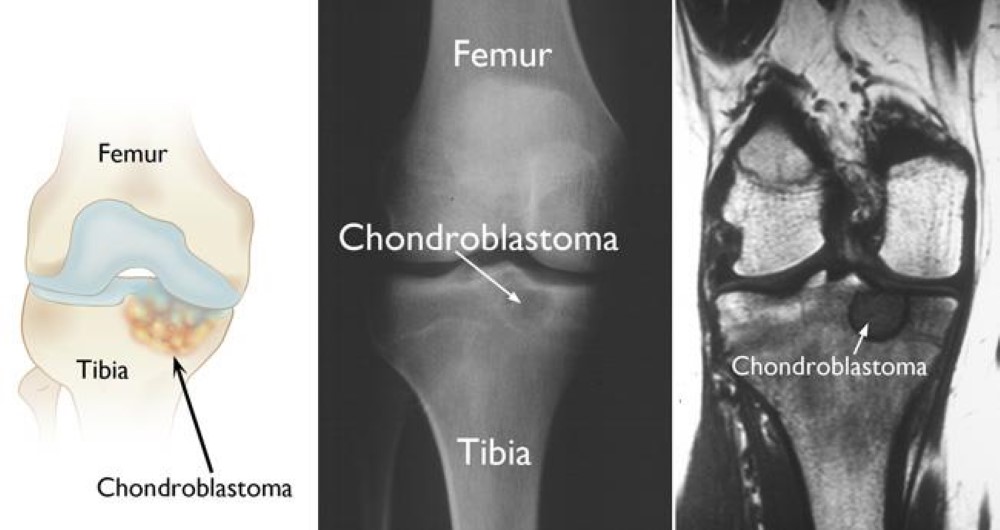
5. Morell, N.; Rajani, R. Chondroblastoma - OrthoInfo - AAOS. https://orthoinfo.aaos.org/en/diseases--conditions/chondroblastoma (accessed Nov 16, 2020).
6.panelRuiGuo111LijuanZheng111Juw WonPark2RuituLv1HaoChen1FangfangJiao1WenqiXu1ShirongMu3HongWen45JinsongQiu6ZhentianWang1PengyuanYang1FeizhenWu1JingyiHui3XiangdongFu6XiaobingShi4512Yujiang GenoShi7812YiXing212…YangShi891012, A. links open overlay; RuiGuo111; 1; 11; LijuanZheng111; Juw WonPark2; 2; RuituLv1; HaoChen1; FangfangJiao1; WenqiXu1; ShirongMu3; 3; HongWen45; 4; 5; JinsongQiu6; 6; ZhentianWang1; PengyuanYang1; FeizhenWu1; JingyiHui3; XiangdongFu6; XiaobingShi4512; 12; Yujiang GenoShi7812; 7; 8; YiXing212; YangShi891012; 9; 10; Highlights•BS69/ZMYND11 binds H3.3K36me3 and colocalizes with H3.3K36me3 in gene bodies•BS69 directly interacts with EFTUD2; SummaryBS69 (also called ZMYND11) contains tandemly arranged PHD. BS69/ZMYND11 Reads and Connects Histone H3.3 Lysine 36 Trimethylation-Decorated Chromatin to Regulated Pre-mRNA Processing. https://reader.elsevier.com/reader/sd/pii/S1097276514006777?token=A4FD3B8CDE2F310EA514C66E96DC4489F79C8EA96F6FC878DCD4BFC066FA809C2E83C8A9B57353A53915171AD2491D4C (accessed Nov 16, 2020).
7. UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. Histone H3.3. https://www.uniprot.org/uniprot/P84243 (accessed Nov 1, 2020).
8.Yuen, B. T. K.; Knoepfler, P. S. Histone H3.3 mutations: a variant path to cancer. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3882088/ (accessed Nov 16, 2020).