1d5r
From Proteopedia
| Line 1: | Line 1: | ||
[[Image:1d5r.gif|left|200px]] | [[Image:1d5r.gif|left|200px]] | ||
| - | + | <!-- | |
| - | + | The line below this paragraph, containing "STRUCTURE_1d5r", creates the "Structure Box" on the page. | |
| - | + | You may change the PDB parameter (which sets the PDB file loaded into the applet) | |
| - | + | or the SCENE parameter (which sets the initial scene displayed when the page is loaded), | |
| - | + | or leave the SCENE parameter empty for the default display. | |
| - | | | + | --> |
| - | | | + | {{STRUCTURE_1d5r| PDB=1d5r | SCENE= }} |
| - | + | ||
| - | + | ||
| - | }} | + | |
'''CRYSTAL STRUCTURE OF THE PTEN TUMOR SUPPRESSOR''' | '''CRYSTAL STRUCTURE OF THE PTEN TUMOR SUPPRESSOR''' | ||
| Line 31: | Line 28: | ||
[[Category: Pavletich, N P.]] | [[Category: Pavletich, N P.]] | ||
[[Category: Yang, H.]] | [[Category: Yang, H.]] | ||
| - | [[Category: | + | [[Category: C2 domain]] |
| - | [[Category: | + | [[Category: Phosphotase]] |
| - | [[Category: | + | [[Category: Phosphotidylinositol]] |
| - | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Fri May 2 13:28:49 2008'' | |
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | |
Revision as of 10:28, 2 May 2008
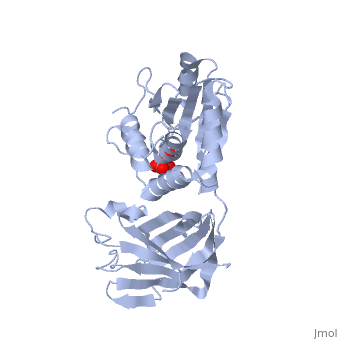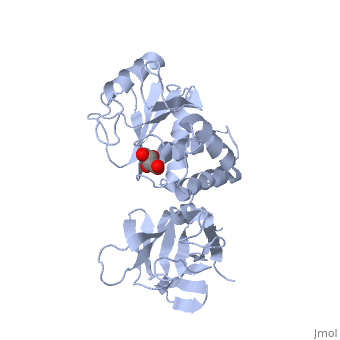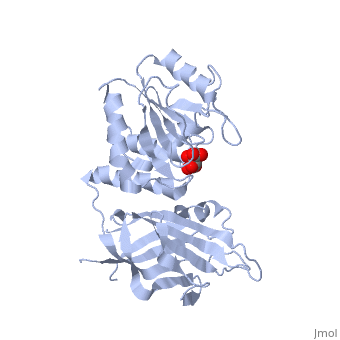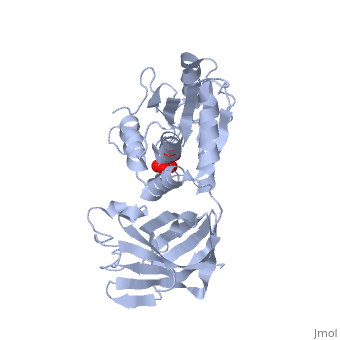
CRYSTAL STRUCTURE OF THE PTEN TUMOR SUPPRESSOR
Overview
The PTEN tumor suppressor is mutated in diverse human cancers and in hereditary cancer predisposition syndromes. PTEN is a phosphatase that can act on both polypeptide and phosphoinositide substrates in vitro. The PTEN structure reveals a phosphatase domain that is similar to protein phosphatases but has an enlarged active site important for the accommodation of the phosphoinositide substrate. The structure also reveals that PTEN has a C2 domain. The PTEN C2 domain binds phospholipid membranes in vitro, and mutation of basic residues that could mediate this reduces PTEN's membrane affinity and its ability to suppress the growth of glioblastoma tumor cells. The phosphatase and C2 domains associate across an extensive interface, suggesting that the C2 domain may serve to productively position the catalytic domain on the membrane.
About this Structure
1D5R is a Single protein structure of sequence from Homo sapiens. Full crystallographic information is available from OCA.
Reference
Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association., Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP, Cell. 1999 Oct 29;99(3):323-34. PMID:10555148 Page seeded by OCA on Fri May 2 13:28:49 2008