Tetrahydroprotoberberine N-methyltransferase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
<StructureSection load='6P3N' size='340' side='right' caption='Tetrahydroprotoberbine complex with SAM (PDB code [[6p3n]])' scene=''> | <StructureSection load='6P3N' size='340' side='right' caption='Tetrahydroprotoberbine complex with SAM (PDB code [[6p3n]])' scene=''> | ||
= Introduction = | = Introduction = | ||
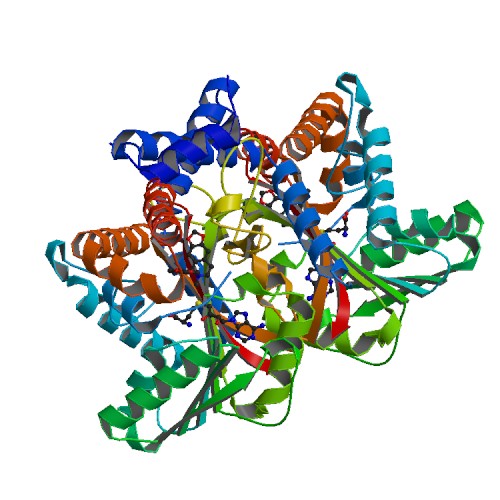
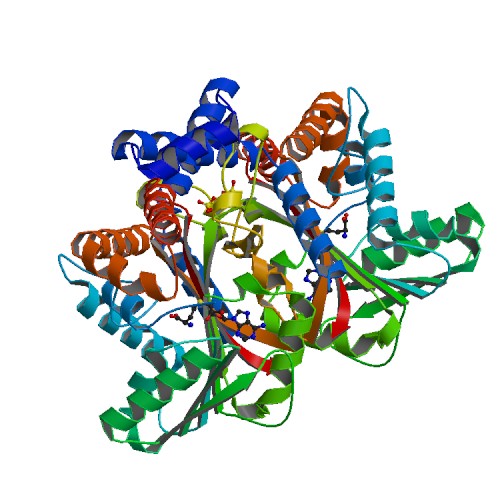
| - | ''' | + | '''Tetrahydroprotoberberine N-methyltransferase''' is a protein whose dimer interface include 6 salt bridges and 8 hydrogens bonds. It is expressed in E. ''coli'' and crystallized at a pH of 7.0. The crystals were grown in the presence of SAH,SAM, and SAH+SMS. Below are the two different substrates that were in the presence of the crystallized protein. The substrate SAM is shown to the right. |
[[Image:6p3o.pdb1-500.jpg]] | [[Image:6p3o.pdb1-500.jpg]] | ||
[[Image:6p3m.pdb1-500.jpg]] | [[Image:6p3m.pdb1-500.jpg]] | ||
= Function = | = Function = | ||
| - | The protein being studied, | + | The protein being studied, Tetrahydroprotoberberine N-methyltransferase, is found in yellow horned poppy (''Glaucium Flavum''). The function of the protein is substate recognition as well as catalysis for the ration engineering of enyzmes for chemoenzymatic synthesis and metabolic engineering. The relative activity of about 8 substrates were tested wiht Protoberberine having the highest percentage. GfTNMT's activity depends on temperature and pH. When the enzyme's activity was at a pH of 8, dropped 10% in activity. 10% activity was also dropped when the temperature was at 30 degrees Celsius. When at 4 degrees Celsius, the activity dropped even more down to 40%. |
= Mutants = | = Mutants = | ||
In TMNT, three amino acid residues in the alpha14-helix form one side of the <scene name='82/829888/Binding_pocket/1'>binding pocket</scene> defining the BP region. The binding pocket consists of His-328(green), Ile-329(purple), and Phe-332(orange). The H328 mutation decreases in activity with stylopine and scoulerine producing a 5- and 2-fold while the activity with THP increases 2-fold. | In TMNT, three amino acid residues in the alpha14-helix form one side of the <scene name='82/829888/Binding_pocket/1'>binding pocket</scene> defining the BP region. The binding pocket consists of His-328(green), Ile-329(purple), and Phe-332(orange). The H328 mutation decreases in activity with stylopine and scoulerine producing a 5- and 2-fold while the activity with THP increases 2-fold. | ||
= Relevance = | = Relevance = | ||
| - | Studying | + | Studying Tetrahydroprotoberberine will provide commercial application where one will gain a lot of knowledge from both the research paper and online sources. Studying this protein will allow readers to engage in the material and apply their own knowledge to better understand the study. This research will provide descriptive roles that TNMT plays such as pathway leading to the formation of different substrates including protoberberine. <ref> Takao, N., Kamigauchi, M., and Okada, M. (1983) Biosynthesis of benzo-[c]phenanthridine alkaloids sanguinarine, chelirubine and macarpine.Helv. Chim. Acta 66, 473–484 CrossRef </ref> |
= Structural Highlights = | = Structural Highlights = | ||
This protein has a <scene name='82/829888/Catalytic_triad/7'>catalytic triad</scene> which consists of amino acids His-208, Glu-204, and Glu-207. The authors explained within the paper that other amino acids may play a role in the triad as well. They were unsure but those three were the most accurate. These amino acids play an important role in catalysis for the protein. <ref> Bennett, M. R., Thompson, M. L., Shepherd, S. A., Dunstan, M. S., Herbert, A. J., Smith, D. R. M., Cronin, V. A., Menon, B. R. K., Levy, C., and Micklefield, J. (2018) Structure and biocatalytic scope of coclaurine Nmethyltransferase.Angew. Chem. Int. Ed. Engl. 57, 10600–10604CrossRef | This protein has a <scene name='82/829888/Catalytic_triad/7'>catalytic triad</scene> which consists of amino acids His-208, Glu-204, and Glu-207. The authors explained within the paper that other amino acids may play a role in the triad as well. They were unsure but those three were the most accurate. These amino acids play an important role in catalysis for the protein. <ref> Bennett, M. R., Thompson, M. L., Shepherd, S. A., Dunstan, M. S., Herbert, A. J., Smith, D. R. M., Cronin, V. A., Menon, B. R. K., Levy, C., and Micklefield, J. (2018) Structure and biocatalytic scope of coclaurine Nmethyltransferase.Angew. Chem. Int. Ed. Engl. 57, 10600–10604CrossRef | ||
| Line 16: | Line 16: | ||
</StructureSection> | </StructureSection> | ||
| - | ==3D structures of | + | ==3D structures of tetrahydroprotoberberine N-methyltransferase== |
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
Revision as of 07:57, 25 November 2020
| |||||||||||
3D structures of tetrahydroprotoberberine N-methyltransferase
Updated on 25-November-2020
6p3m – GfTHPBM + SAH – Glaucum flavum
6p3n – GfTHPBM + SAM
6p3n – GfTHPBM + SAH + methylstylopine
References
- ↑ Takao, N., Kamigauchi, M., and Okada, M. (1983) Biosynthesis of benzo-[c]phenanthridine alkaloids sanguinarine, chelirubine and macarpine.Helv. Chim. Acta 66, 473–484 CrossRef
- ↑ Bennett, M. R., Thompson, M. L., Shepherd, S. A., Dunstan, M. S., Herbert, A. J., Smith, D. R. M., Cronin, V. A., Menon, B. R. K., Levy, C., and Micklefield, J. (2018) Structure and biocatalytic scope of coclaurine Nmethyltransferase.Angew. Chem. Int. Ed. Engl. 57, 10600–10604CrossRef Medline