We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:R. Jeremy Johnson/Mitochondrial Calcium Uniporter
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
The mitochondrial calcium uniporter (MCU) complex is the main source of entry for [https://en.wikipedia.org/wiki/Calcium calcium] ions into the [https://en.wikipedia.org/wiki/Mitochondrial_matrix mitochondrial matrix] from the [https://en.wikipedia.org/wiki/Mitochondrion#Intermembrane_space intermembrane space]. MCU channels exist in most [https://en.wikipedia.org/wiki/Eukaryote eukaryotic] life, but activity is regulated differently in each [https://en.wikipedia.org/wiki/Clade clade].<ref name="Baradaran">PMID:29995857</ref> The precise identity of the MCU wasn't discovered until 2011 and was discovered using a combination of [https://en.wikipedia.org/wiki/Nuclear_magnetic_resonance_spectroscopy NMR spectroscopy], [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryoelectron microscopy], and [https://en.wikipedia.org/wiki/X-ray_crystallography x-ray crystallography].<ref name="Woods">PMID:31869674</ref> [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryoelectron microscopy] (Cryo-EM) was instrumental in elucidating the complete structure of the MCU complex, which subsequently provides a structural framework for understanding the mechanism by which the MCU functions.<ref name="Giorgi" /> Prior modeling of the structure was difficult because it has no apparent sequence similarity to other ion channels.<ref name="Baradaran"/> However, like other ion channels, the MCU is highly selective and efficient. The MCU has the ability to only allow calcium ions into the mitochondrial matrix at a rate of 5,000,000 ions per second, even though [https://en.wikipedia.org/wiki/Potassium potassium] ions are over 100,000 times more abundant in the intermembrane space.<ref name="Baradaran"/> | The mitochondrial calcium uniporter (MCU) complex is the main source of entry for [https://en.wikipedia.org/wiki/Calcium calcium] ions into the [https://en.wikipedia.org/wiki/Mitochondrial_matrix mitochondrial matrix] from the [https://en.wikipedia.org/wiki/Mitochondrion#Intermembrane_space intermembrane space]. MCU channels exist in most [https://en.wikipedia.org/wiki/Eukaryote eukaryotic] life, but activity is regulated differently in each [https://en.wikipedia.org/wiki/Clade clade].<ref name="Baradaran">PMID:29995857</ref> The precise identity of the MCU wasn't discovered until 2011 and was discovered using a combination of [https://en.wikipedia.org/wiki/Nuclear_magnetic_resonance_spectroscopy NMR spectroscopy], [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryoelectron microscopy], and [https://en.wikipedia.org/wiki/X-ray_crystallography x-ray crystallography].<ref name="Woods">PMID:31869674</ref> [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryoelectron microscopy] (Cryo-EM) was instrumental in elucidating the complete structure of the MCU complex, which subsequently provides a structural framework for understanding the mechanism by which the MCU functions.<ref name="Giorgi" /> Prior modeling of the structure was difficult because it has no apparent sequence similarity to other ion channels.<ref name="Baradaran"/> However, like other ion channels, the MCU is highly selective and efficient. The MCU has the ability to only allow calcium ions into the mitochondrial matrix at a rate of 5,000,000 ions per second, even though [https://en.wikipedia.org/wiki/Potassium potassium] ions are over 100,000 times more abundant in the intermembrane space.<ref name="Baradaran"/> | ||
| - | Under resting conditions, the calcium concentration in the mitochondria is about the same as in the [https://en.wikipedia.org/wiki/Cytoplasm cytoplasm], but when stimulated, it can increase calcium concentration 10 to 20-fold.<ref name="Giorgi">PMID:30143745</ref> | + | Under resting conditions, the calcium concentration in the mitochondria is about the same as in the [https://en.wikipedia.org/wiki/Cytoplasm cytoplasm], but when stimulated, it can increase calcium concentration 10 to 20-fold.<ref name="Giorgi">PMID:30143745</ref> ([https://en.wikipedia.org/wiki/Mitochondria_associated_membranes Mitochondria-associated ER membranes]) exist between mitochondria and the [https://en.wikipedia.org/wiki/Endoplasmic_reticulum endoplasmic reticulum], the two largest cellular stores of calcium, to facilitate efficient transport of calcium ions.<ref name="Wang">PMID:28882140</ref> The transfer of electrons through [https://en.wikipedia.org/wiki/Electron_transport_chain#Mitochondrial_redox_carriers respiratory complexes I-IV] produces the energy to pump [https://en.wikipedia.org/wiki/Hydrogen_ion hydrogen ions] into the intermembrane space and create the proton [https://en.wikipedia.org/wiki/Electrochemical_gradient electrochemical gradient] potential.<ref name="Giorgi"/> This negative electrochemical potential is the driving force that moves positively charged calcium ions into the mitochondrial matrix.<ref name="Giorgi"/> |
Regulation of the uptake and efflux of calcium is important to increase calcium levels enough to activate certain enzymes, but also avoid calcium overload and [https://en.wikipedia.org/wiki/Apoptosis apoptosis].<ref name="Wang"/> Mitochondrial calcium increases [http://proteopedia.org/wiki/index.php/ATP ATP] production by activating [http://proteopedia.org/wiki/index.php/Pyruvate_dehydrogenase pyruvate dehydrogenase], [https://en.wikipedia.org/wiki/Oxoglutarate_dehydrogenase_complex α-ketoglutarate dehydrogenase], and [http://proteopedia.org/wiki/index.php/Isocitrate_dehydrogenase isocitrate dehydrogenase] in the [https://en.wikipedia.org/wiki/Citric_acid_cycle Krebs cycle].<ref name="Wang"/> Therefore, a deficiency of MCU leads to decreases in associated enzyme activity and of [https://en.wikipedia.org/wiki/Oxidative_phosphorylation oxidative phosphorylation]. | Regulation of the uptake and efflux of calcium is important to increase calcium levels enough to activate certain enzymes, but also avoid calcium overload and [https://en.wikipedia.org/wiki/Apoptosis apoptosis].<ref name="Wang"/> Mitochondrial calcium increases [http://proteopedia.org/wiki/index.php/ATP ATP] production by activating [http://proteopedia.org/wiki/index.php/Pyruvate_dehydrogenase pyruvate dehydrogenase], [https://en.wikipedia.org/wiki/Oxoglutarate_dehydrogenase_complex α-ketoglutarate dehydrogenase], and [http://proteopedia.org/wiki/index.php/Isocitrate_dehydrogenase isocitrate dehydrogenase] in the [https://en.wikipedia.org/wiki/Citric_acid_cycle Krebs cycle].<ref name="Wang"/> Therefore, a deficiency of MCU leads to decreases in associated enzyme activity and of [https://en.wikipedia.org/wiki/Oxidative_phosphorylation oxidative phosphorylation]. | ||
==Mitochondrial Calcium Uniporter Complex== | ==Mitochondrial Calcium Uniporter Complex== | ||
| - | [[Image:structure.png|300 px|right|thumb|Figure 1: Structure of mitochondrial calcium uniporter colored by functional domain. The transmembrane domain is highlighted in salmon, the matrix domain in light cyan, coiled-coil domain in dark violet, and the N-terminal domain in slate blue. [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] | ||
The mitochondrial calcium uniporter complex exists as a large complex (around 480 kDa in humans) made up of both pore-forming and regulatory subunits.<ref name="Wang"/> The MCU is a complex composed of regulatory subunits including mitochondrial calcium uptake (MICU), essential MICU regulator (EMRE), MCU regulatory subunit b (MCUb), and MCU regulator 1 (MCUR1). <ref name="Fan" /> The mitochondrial uptake proteins (MICU1 and MICU2) are regulatory proteins in the MCU complex that exist in the intermembrane space and contain [https://en.wikipedia.org/wiki/EF_hand EF hand domains] for calcium binding to control transport through the channel of the MCU complex.<ref name="Wang"/> When calcium ion concentration in the intermembrane space is low, MICU1 and 2 block the MCU to prevent uptake of calcium.<ref name="Wang"/> In the presence of high calcium concentrations, more calcium binds to these regulatory proteins and they undergo a conformational change to allow calcium ions through the MCU and into the matrix.<ref name="Wang"/> In fact, when calcium levels are below 500 nM, MICU1 can block movement of calcium by itself, calcium levels between 500 nM and 1,500 nM require both MICU1 and MICU2 to block ion entry, and any concentration over 1,500 nM is sufficient for calcium entry.<ref name="Giorgi"/> Another regulatory protein, MCUR1 is a cofactor in the assembly of the [https://en.wikipedia.org/wiki/Electron_transport_chain respiratory chain] rather than an essential part of the uniporter.<ref name="Giorgi"/> Though the MCU is able to take up calcium independently, there are two other pore-forming subunits, the MCUb and the essential MCU regulator (EMRE).<ref name="Wang"/> MCUb is similar to MCU, but certain amino acids differ and make it an inhibitory subunit.<ref name="Wang"/> The EMRE is located in the intermembrane space and connects MICU1 and MICU2 to the MCU.<ref name="Giorgi"/> It also contributes to regulation of calcium intake in the MCU.<ref name="Wang"/> | The mitochondrial calcium uniporter complex exists as a large complex (around 480 kDa in humans) made up of both pore-forming and regulatory subunits.<ref name="Wang"/> The MCU is a complex composed of regulatory subunits including mitochondrial calcium uptake (MICU), essential MICU regulator (EMRE), MCU regulatory subunit b (MCUb), and MCU regulator 1 (MCUR1). <ref name="Fan" /> The mitochondrial uptake proteins (MICU1 and MICU2) are regulatory proteins in the MCU complex that exist in the intermembrane space and contain [https://en.wikipedia.org/wiki/EF_hand EF hand domains] for calcium binding to control transport through the channel of the MCU complex.<ref name="Wang"/> When calcium ion concentration in the intermembrane space is low, MICU1 and 2 block the MCU to prevent uptake of calcium.<ref name="Wang"/> In the presence of high calcium concentrations, more calcium binds to these regulatory proteins and they undergo a conformational change to allow calcium ions through the MCU and into the matrix.<ref name="Wang"/> In fact, when calcium levels are below 500 nM, MICU1 can block movement of calcium by itself, calcium levels between 500 nM and 1,500 nM require both MICU1 and MICU2 to block ion entry, and any concentration over 1,500 nM is sufficient for calcium entry.<ref name="Giorgi"/> Another regulatory protein, MCUR1 is a cofactor in the assembly of the [https://en.wikipedia.org/wiki/Electron_transport_chain respiratory chain] rather than an essential part of the uniporter.<ref name="Giorgi"/> Though the MCU is able to take up calcium independently, there are two other pore-forming subunits, the MCUb and the essential MCU regulator (EMRE).<ref name="Wang"/> MCUb is similar to MCU, but certain amino acids differ and make it an inhibitory subunit.<ref name="Wang"/> The EMRE is located in the intermembrane space and connects MICU1 and MICU2 to the MCU.<ref name="Giorgi"/> It also contributes to regulation of calcium intake in the MCU.<ref name="Wang"/> | ||
| + | [[Image:structure.png|300 px|right|thumb|Figure 1: Structure of mitochondrial calcium uniporter colored by functional domain. The transmembrane domain is highlighted in salmon, the matrix domain in light cyan, coiled-coil domain in dark violet, and the N-terminal domain in slate blue. [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] | ||
==MCU Structure== | ==MCU Structure== | ||
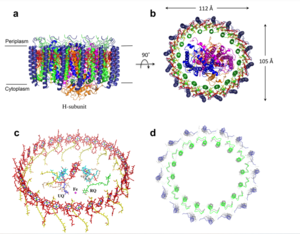
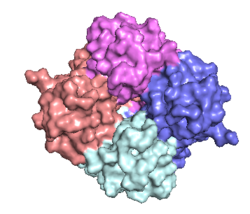
| - | The <scene name='83/832952/Starting_scene/5'>mitochondrial calcium uniporter (MCU)</scene> is the ion channel component (Figure 1). The MCU was originally thought to | + | The <scene name='83/832952/Starting_scene/5'>mitochondrial calcium uniporter (MCU)</scene> is the ion channel component of the complex (Figure 1). The MCU was originally thought to a pentamer of five identical subunits, but based on the structure was shown to exist as four monomers, identical in sequence, arranged and packed together such that they structurally form a <scene name='83/832952/Dimer_of_dimers/5'>dimer of dimers</scene> (Figure 2).<ref name="Woods">PMID:31869674</ref> The MCU protein is composed of a <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene>, a <scene name='83/837230/Coiled_coil/3'>coiled coil domain</scene>, and a <scene name='83/837230/Nterm/2'>N-Terminal Domain</scene> (NTD) (Figure 1).<ref name="Woods"/> The hydrophobic <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene> is located in the ([https://en.wikipedia.org/wiki/Inner_mitochondrial_membrane inner mitochondrial membrane]) and the hydrophilic coiled-coil domain exists in the mitochondrial matrix.<ref name="Baradaran"/> |
[[Image:Nterm.png|250 px|right|thumb|Figure 2: Symmetry and organization of subunits from looking down into the uniporter from the inner mitochondrial membrane [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] | [[Image:Nterm.png|250 px|right|thumb|Figure 2: Symmetry and organization of subunits from looking down into the uniporter from the inner mitochondrial membrane [https://en.wikipedia.org/wiki/Protein_Data_Bank PDB] [https://www.rcsb.org/structure/6DT0 6DT0]]] | ||
===Transmembrane Domain=== | ===Transmembrane Domain=== | ||
| - | The <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene> is on the | + | The <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene> is on the inner mitochondrial membrane open to the inner membrane space (Figure 1). The <scene name='83/837230/Transmembrane_domain/3'>transmembrane domain</scene> consists of eight separate helices (TM1 and TM2 from each subunit) that are connected by mostly hydrophobic amino acids in the intermembrane space and has <scene name='83/832952/Starting_scene/5'>four-fold</scene> symmetry (Figure 2).<ref name="Baradaran"/> The small <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> pore, highly specific for calcium binding is located in the interior of the bundle of four packed <scene name='83/832952/Tm2/2'>TM2</scene> helices while the four helices of <scene name='83/837230/Transmembrane_1/2'>TM1</scene> surround this TM2 bundle. <scene name='83/837230/Transmembrane_1/2'>TM1</scene> packs tightly against <scene name='83/832952/Tm2/2'>TM2</scene> from the neighboring subunit which conveys a sense of domain-swapping.<ref name="Fan">PMID:29995856</ref> |
| - | ===Coiled-coil | + | ===Coiled-coil Domain=== |
Past the transmembrane domain, the N-terminal domains of the <scene name='83/832952/Tm1/2'>TM1</scene> helices extend into the matrix and form coiled-coils with a C-terminal helix.<ref name="Baradaran"/> These "legs" are separated from each other which allows enough space for calcium ions to diffuse out into the matrix.<ref name="Baradaran"/> The <scene name='83/837230/Coiled_coil/3'>coiled coil</scene> domain is the first subsection of the soluble domain, which resides in the inner mitochondrial membrane. The coiled coil functions as the joints of the uniporter, providing flexibility to promote transport of Ca<sup>2+</sup>ions down their concentration gradient.<ref name="Fan" /> When Ca<sup>2+</sup> ions binds to the selectivity pore, the coiled-coil swings approximately 8° around its end near the <scene name='83/837230/Nterm/2'>NTD</scene>. This movement propagates to the top of the transmembrane domain, where the pore is located about 85 Å away. The largest displacement triggered by the movement of the coiled-coil is in the transmembrane domain, where the coil bends 20°, moving the transmembrane domain further apart. The junction between the transmembrane domain and the coiled coil's flexibility can be attributed to the disordered packing between subunits. Subunits A and C adopt different conformations than the B and D subunits, although they superimpose closely.<ref name="Fan" /> The coiled-coil domain is also responsible for assembly of the MCU and is [https://en.wikipedia.org/wiki/Post-translational_modification post-translationally modified].<ref name="Fan"/> | Past the transmembrane domain, the N-terminal domains of the <scene name='83/832952/Tm1/2'>TM1</scene> helices extend into the matrix and form coiled-coils with a C-terminal helix.<ref name="Baradaran"/> These "legs" are separated from each other which allows enough space for calcium ions to diffuse out into the matrix.<ref name="Baradaran"/> The <scene name='83/837230/Coiled_coil/3'>coiled coil</scene> domain is the first subsection of the soluble domain, which resides in the inner mitochondrial membrane. The coiled coil functions as the joints of the uniporter, providing flexibility to promote transport of Ca<sup>2+</sup>ions down their concentration gradient.<ref name="Fan" /> When Ca<sup>2+</sup> ions binds to the selectivity pore, the coiled-coil swings approximately 8° around its end near the <scene name='83/837230/Nterm/2'>NTD</scene>. This movement propagates to the top of the transmembrane domain, where the pore is located about 85 Å away. The largest displacement triggered by the movement of the coiled-coil is in the transmembrane domain, where the coil bends 20°, moving the transmembrane domain further apart. The junction between the transmembrane domain and the coiled coil's flexibility can be attributed to the disordered packing between subunits. Subunits A and C adopt different conformations than the B and D subunits, although they superimpose closely.<ref name="Fan" /> The coiled-coil domain is also responsible for assembly of the MCU and is [https://en.wikipedia.org/wiki/Post-translational_modification post-translationally modified].<ref name="Fan"/> | ||
| Line 39: | Line 39: | ||
===Mutations=== | ===Mutations=== | ||
| - | A number of mutations completely eliminate calcium uptake by the MCU. For example, mutation of | + | A number of mutations completely eliminate calcium uptake by the MCU. For example, mutation of Trp224, Asp225, Glu228, or Pro229 of the <scene name='83/832952/Dxxe_motif/7'>WDXXEP motif</scene> altered the highly conserved selectivity filter and completely eliminated calcium uptake.<ref name="Baradaran"/><ref name="Fan"/> Even mutating Glu228 to an aspartate significantly changes the dimensions of the pore and inhibits uptake of calcium. However, mutation of either X residue was not detrimental to calcium uptake.<ref name="Baradaran"/> Furthermore, mutation of the tyrosine residue directly below the selectivity filter substantially impaired calcium intake and proper protein folding.<ref name="Fan"/> The residue on TM1 that affected calcium uptake the most in human MCU was Trp317 (analogous to <scene name='83/832952/New_ones/7'>Trp210</scene> in ''C. europaea'') which has a side chain constituting a primary contact point between TM1 and TM2.<ref name="Fan"/> Mutation of human MCU Phe326 (analogous to <scene name='83/832952/New_ones/8'>Phe218</scene> in ''C. europaea'') or Gly331 of the TM1-TM2 linker (<scene name='83/832952/New_ones/9'>Gly223</scene> in ''C. europaea'') affected the linker conformation and configuration of the pore entrance and impaired calcium intake.<ref name="Fan"/> |
==Regulation and Inhibition== | ==Regulation and Inhibition== | ||
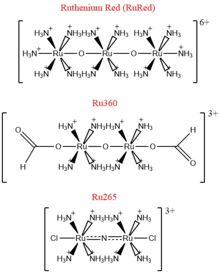
[[Image:Ruthenium_Inhibitors.jpg|220 px|right|thumb|Figure 4: Structures of the ruthenium-based inhibitors of the MCU. Created using ChemDraw Professional 16.0]] | [[Image:Ruthenium_Inhibitors.jpg|220 px|right|thumb|Figure 4: Structures of the ruthenium-based inhibitors of the MCU. Created using ChemDraw Professional 16.0]] | ||
| - | The most well-known and commonly used inhibitor of the MCU is [https://en.wikipedia.org/wiki/Ruthenium_red ruthenium red] (RuRed).<ref name="Woods"/> RuRed is a trinuclear, oxo-bridged complex that effectively inhibits calcium uptake without affecting mitochondrial respiration or calcium efflux. | + | The most well-known and commonly used inhibitor of the MCU is [https://en.wikipedia.org/wiki/Ruthenium_red ruthenium red] (RuRed).<ref name="Woods"/> RuRed is a trinuclear, oxo-bridged complex that effectively inhibits calcium uptake without affecting mitochondrial respiration or calcium efflux. The disadvantage of ruthenium red is its challenging purification.<ref name="Woods"/> Interestingly, a compound identified in an impure version of RuRed, termed [https://en.wikipedia.org/wiki/Ru360 Ru360], was found to be the actual active component as an inhibitor of the MCU (Figure 4). Ru360 is a binuclear, oxo-bridged complex with a similar structure to that of RuRed. The only flaw with Ru360 was that it showed low cell permeability, so Ru265 was subsequently developed with twice the cell permeability of Ru360. Ru265 possesses two bridged Ru centers bridged by a nitride ligand (Figure 4).<ref name="Woods"/> |
| - | Recent experiments suggest that Ru360 inhibits calcium uptake through interactions with the <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> motif. | + | Recent experiments suggest that Ru360 inhibits calcium uptake through interactions with the <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> motif. However, not much is known about the mode of inhibition. Also, mutations of Asp261 and Ser259 in human MCU were shown to maintain calcium uptake into the matrix, but reduce the inhibitory effect of Ru360, but not Ru265. However, a mutation in a cysteine residue in the <scene name='83/832952/New_ones/4'>NTD</scene> had the opposite effect as it reduced the inhibitory effects of Ru265, but not Ru360 (Figure 4).<ref name="Woods"/> |
==Medical Relevance== | ==Medical Relevance== | ||
| - | The MCU has a large role in disease because of its effect on apoptosis and cell signaling. The overload of the mitochondrial matrix with calcium leads to release of [https://en.wikipedia.org/wiki/Cytochrome_c cytochrome c], overproduction of [https://en.wikipedia.org/wiki/Reactive_oxygen_species reactive oxygen species], mitochondrial swelling, and the opening of the mitochondrial permeability transition pore ([https://en.wikipedia.org/wiki/Mitochondrial_permeability_transition_pore mPTP]) which all lead to apoptotic cell death.<ref name="Woods"/> This connection between mitochondrial calcium and apoptosis makes | + | The MCU has a large role in disease because of its effect on apoptosis and cell signaling. The overload of the mitochondrial matrix with calcium leads to release of [https://en.wikipedia.org/wiki/Cytochrome_c cytochrome c], overproduction of [https://en.wikipedia.org/wiki/Reactive_oxygen_species reactive oxygen species], mitochondrial swelling, and the opening of the mitochondrial permeability transition pore ([https://en.wikipedia.org/wiki/Mitochondrial_permeability_transition_pore mPTP]) which all lead to apoptotic cell death.<ref name="Woods"/> This connection between mitochondrial calcium and apoptosis makes MCU dysregulation a large contributor to cell death and disease. Calcium machinery in the mitochondria are targets for [https://en.wikipedia.org/wiki/Oncogene proto-oncogenes] and [https://en.wikipedia.org/wiki/Tumor_suppressor tumor suppressors] for this very reason.<ref name="Giorgi"/> Apoptosis can either be induced or repressed. Furthermore, external stimuli can activate receptors in the endoplasmic reticulum that release calcium and activate signal transductions.<ref name="Wang"/> Sequestration of calcium in the mitochondria is vital to shut down these activations, so any impact in movement of calcium ions can cause a wide variety of diseases.<ref name="Wang"/> |
===Neurodegenerative Disorders=== | ===Neurodegenerative Disorders=== | ||
| - | Disruption in calcium homeostasis leads to a wide range of [https://en.wikipedia.org/wiki/Neurodegeneration neurodegenerative disorders]. The MCU complex has been identified to play a large role in [https://en.wikipedia.org/wiki/Neuromuscular_disease neuromuscular disease] because of a loss | + | Disruption in calcium homeostasis leads to a wide range of [https://en.wikipedia.org/wiki/Neurodegeneration neurodegenerative disorders]. The MCU complex has been identified to play a large role in [https://en.wikipedia.org/wiki/Neuromuscular_disease neuromuscular disease] because of a loss of function of the MICU1 subunit.<ref name="Woods"/> This causes [https://en.wikipedia.org/wiki/Myopathy myopathy], learning difficulties, and progressive movement disorders which can be lethal. In [https://en.wikipedia.org/wiki/Alzheimer%27s_disease Alzheimer's disease], the buildup of [https://en.wikipedia.org/wiki/Amyloid_beta amyloid-β] plaques in the brain leads to increased calcium uptake in [https://en.wikipedia.org/wiki/Neuron neurons] and cell death. Similarly, in early onset [https://en.wikipedia.org/wiki/Parkinson%27s_disease Parkinson's disease], degradation of MICU1 by the ligase [http://proteopedia.org/wiki/index.php/Parkin Parkin] leads to increased mitochondrial calcium uptake, overload, and death. Finally, disrupted glutamate homeostasis in [https://en.wikipedia.org/wiki/Astrocyte astrocytes] and neurons leads to calcium overload and cell death via [https://en.wikipedia.org/wiki/Excitotoxicity excitotoxicity] in Amyotrophic Lateral Sclerosis ([https://en.wikipedia.org/wiki/Amyotrophic_lateral_sclerosis ALS]).<ref name="Woods"/> |
===Diabetes=== | ===Diabetes=== | ||
| - | Calcium homeostasis misregulation has also been proven to be instrumental in [https://en.wikipedia.org/wiki/Obesity obesity], [https://en.wikipedia.org/wiki/Insulin_resistance insulin resistance], and [https://en.wikipedia.org/wiki/Type_2_diabetes type-II diabetes].<ref name="Wang"/> The intracellular calcium concentrations in primary [https://en.wikipedia.org/wiki/Adipocyte adipocytes] from obese human subjects has been found to be elevated. | + | Calcium homeostasis misregulation has also been proven to be instrumental in [https://en.wikipedia.org/wiki/Obesity obesity], [https://en.wikipedia.org/wiki/Insulin_resistance insulin resistance], and [https://en.wikipedia.org/wiki/Type_2_diabetes type-II diabetes].<ref name="Wang"/> The intracellular calcium concentrations in primary [https://en.wikipedia.org/wiki/Adipocyte adipocytes] from obese human subjects has been found to be elevated. Any inhibition of downstream calcium signaling could decrease movement of the [http://proteopedia.org/wiki/index.php/GLUT4 GLUT4] glucose transporter and glucose uptake. Additionally, removal of MCU in [https://en.wikipedia.org/wiki/Beta_cell β-cells] in the [https://en.wikipedia.org/wiki/Pancreas pancreas] demonstrated a decrease in cellular ATP concentration following glucose stimulation which resulted in decreased glucose-stimulated insulin secretion. |
===Heart Failure=== | ===Heart Failure=== | ||
| - | Calcium overload in the mitochondria of cardiac cells lead to [https://en.wikipedia.org/wiki/Apoptosis apoptotic] cardiac cell death. Calcium governs [https://en.wikipedia.org/wiki/Cardiac_excitation-contraction_coupling excitation contraction coupling] | + | Calcium overload in the mitochondria of cardiac cells lead to [https://en.wikipedia.org/wiki/Apoptosis apoptotic] cardiac cell death. Calcium governs [https://en.wikipedia.org/wiki/Cardiac_excitation-contraction_coupling excitation contraction coupling] of the cardiac muscles, which creates the ATP needed to power the contraction during heart beats. The increase in mitochondrial Ca<sup>2+</sup> concentration is essential for the functioning of this muscle contraction. Mitochondrial Ca<sup>2+</sup> overload, though, leads to [https://en.wikipedia.org/wiki/Necrosis necrotic] cardiac cell death and can be targeted with regulation of the MCU. An example of potential treatment might involve the use of Ru360 to inhibit the uptake of Ca<sup>2+</sup> ions into the mitochondria.<ref name="Giorgi" /> |
</StructureSection> | </StructureSection> | ||
==References== | ==References== | ||
Revision as of 13:46, 2 December 2020
Mitochondrial Calcium Uniporter
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 1.22 1.23 1.24 1.25 Baradaran R, Wang C, Siliciano AF, Long SB. Cryo-EM structures of fungal and metazoan mitochondrial calcium uniporters. Nature. 2018 Jul 11. pii: 10.1038/s41586-018-0331-8. doi:, 10.1038/s41586-018-0331-8. PMID:29995857 doi:http://dx.doi.org/10.1038/s41586-018-0331-8
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 2.9 Woods JJ, Wilson JJ. Inhibitors of the mitochondrial calcium uniporter for the treatment of disease. Curr Opin Chem Biol. 2019 Dec 20;55:9-18. doi: 10.1016/j.cbpa.2019.11.006. PMID:31869674 doi:http://dx.doi.org/10.1016/j.cbpa.2019.11.006
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Giorgi C, Marchi S, Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol. 2018 Nov;19(11):713-730. doi: 10.1038/s41580-018-0052-8. PMID:30143745 doi:http://dx.doi.org/10.1038/s41580-018-0052-8
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 Wang CH, Wei YH. Role of mitochondrial dysfunction and dysregulation of Ca(2+) homeostasis in the pathophysiology of insulin resistance and type 2 diabetes. J Biomed Sci. 2017 Sep 7;24(1):70. doi: 10.1186/s12929-017-0375-3. PMID:28882140 doi:http://dx.doi.org/10.1186/s12929-017-0375-3
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 Fan C, Fan M, Orlando BJ, Fastman NM, Zhang J, Xu Y, Chambers MG, Xu X, Perry K, Liao M, Feng L. X-ray and cryo-EM structures of the mitochondrial calcium uniporter. Nature. 2018 Jul 11. pii: 10.1038/s41586-018-0330-9. doi:, 10.1038/s41586-018-0330-9. PMID:29995856 doi:http://dx.doi.org/10.1038/s41586-018-0330-9
- ↑ Yoo J, Wu M, Yin Y, Herzik MA Jr, Lander GC, Lee SY. Cryo-EM structure of a mitochondrial calcium uniporter. Science. 2018 Jun 28. pii: science.aar4056. doi: 10.1126/science.aar4056. PMID:29954988 doi:http://dx.doi.org/10.1126/science.aar4056
Student Contributors
Ryan Heumann
Lizzy Ratz
Holly Rowe
Madi Summers
Rieser Wells