We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:R. Jeremy Johnson/Mitochondrial Calcium Uniporter
From Proteopedia
(Difference between revisions)
| Line 28: | Line 28: | ||
The <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> of the MCU is composed of multiple layers of acidic amino acids near the narrow mouth of the channel and is responsible for the high affinity and selectivity of the MCU for calcium ([https://en.wikipedia.org/wiki/Dissociation_constant dissociation constant] of less than 2nM) (Figure 3).<ref name="Baradaran"/> Negatively charged aspartates <scene name='83/832952/New_ones/2'>(Asp333)</scene> at the mouth of the MCU congregate positively charged <scene name='83/832952/Calcium/4'>calcium ions</scene> at the entrance of the channel.<ref name="Baradaran"/> A highly conserved <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> [https://en.wikipedia.org/wiki/Sequence_motif motif] in the TM2 helices form the selectivity pore which selects for calcium transport over other similar ions.<ref name="Baradaran"/> | The <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> of the MCU is composed of multiple layers of acidic amino acids near the narrow mouth of the channel and is responsible for the high affinity and selectivity of the MCU for calcium ([https://en.wikipedia.org/wiki/Dissociation_constant dissociation constant] of less than 2nM) (Figure 3).<ref name="Baradaran"/> Negatively charged aspartates <scene name='83/832952/New_ones/2'>(Asp333)</scene> at the mouth of the MCU congregate positively charged <scene name='83/832952/Calcium/4'>calcium ions</scene> at the entrance of the channel.<ref name="Baradaran"/> A highly conserved <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> [https://en.wikipedia.org/wiki/Sequence_motif motif] in the TM2 helices form the selectivity pore which selects for calcium transport over other similar ions.<ref name="Baradaran"/> | ||
| - | The <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> motif consists of <scene name='83/832952/Tryptophan/2'> | + | The <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> motif consists of <scene name='83/832952/Tryptophan/2'>Trp332</scene> at the N-terminal end, <scene name='83/832952/Selectivity_filter_asp/2'>Asp333</scene>, <scene name='83/832952/Selectivity_filter_glu/3'>Glu336</scene>, and <scene name='83/832952/New_ones/5'>Pro337</scene>.<ref name="Baradaran"/> The negatively charged side chains of Asp333 and <scene name='83/832933/Glu_358/4'>Glu336</scene> point towards the pore.<ref name="Baradaran"/> The <scene name='83/832933/Diameter/2'>diameter</scene> of the pore created by the carboxyl ring on the 4 identical glutamates (Glu336) is about 2.8Å, allowing only a dehydrated Ca<sup>2+</sup> ion to bind. The combination of these radii and high negative charge (Figure 3) account for the selectivity of the MCU. For example, potassium has an [https://en.wikipedia.org/wiki/Ionic_radius ionic radius] of 1.38Å which is much larger than the 1.00Å ionic radius of calcium and thus cannot fit through the pore.<ref name="Baradaran"/> Additionally, even though sodium ions have a similar ionic radius, the +2 charge on calcium is better matched for coordination with the glutamate residues.<ref name="Baradaran"/> |
| - | scene name='83/832952/Tryptophan_proline/2'> | + | The additional residues of the WDXXEP motif, <scene name='83/832952/Tryptophan_proline/2'>Trp332 and Pro337</scene> pack against each other, are oriented towards the pore, and serve to stabilize <scene name='83/832952/Selectivity_filter_glu/4'>Asp333 and Glu336</scene>.<ref name="Baradaran"/><ref name="Fan"/> Trp332 stabilizes the carbonyl side chains of Glu336 through <scene name='83/832933/H_bond_trp354_glu358/3'>hydrogen bonding</scene> and anion pi interactions. Approximately one helical turn below the glutamate ring of the selectivity filter, a wider tyrosine ring (12Å) facilitates calcium rehydration after passage through the selectivity pore.<ref name="Fan"/> |
| - | + | ||
===Movement of Calcium=== | ===Movement of Calcium=== | ||
| - | Cryo-EM showed three <scene name='83/832952/Starting_scene/5'>calciums</scene> in the MCU channel of roughly spherical density equally spaced 6Å apart.<ref name="Baradaran"/> Sites 1 and 2 lie within the <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> and likely contain calcium, but site 3 could be calcium or some other small molecule.<ref name="Baradaran"/> Site 1 is positioned in the ring formed by <scene name='83/832952/Selectivity_filter_asp/2'> | + | Cryo-EM showed three <scene name='83/832952/Starting_scene/5'>calciums</scene> in the MCU channel of roughly spherical density equally spaced 6Å apart.<ref name="Baradaran"/> Sites 1 and 2 lie within the <scene name='83/832952/Selectivity_filter/3'>selectivity filter</scene> and likely contain calcium, but site 3 could be calcium or some other small molecule.<ref name="Baradaran"/> Site 1 is positioned in the ring formed by <scene name='83/832952/Selectivity_filter_asp/2'>Asp333</scene> residues with a distance of 4Å between the center of the site and each [https://en.wikipedia.org/wiki/Carboxylate carboxylate group] indicating the presence of water.<ref name="Baradaran"/> Site 2 is positioned in the ring formed by <scene name='83/832952/Selectivity_filter_glu/3'>Glu336</scene> with a smaller distance (2.8Å) between the carboxylate group of each residue and the middle of the site, indicating the absence of water.<ref name="Baradaran"/> For transporting calcium, a mechanism has been proposed where one calcium ion coordinated with water positioned in site 1 is dehydrated and moves to site 2 while a new calcium ion moves from the intermembrane space into site 1.<ref name="Baradaran"/> Meanwhile, a different calcium ion moves from site 2 to site 3 and becomes rehydrated upon passage into the mitochondrial matrix.<ref name="Baradaran"/> |
===Mutations=== | ===Mutations=== | ||
| - | A number of mutations completely eliminate calcium uptake by the MCU. For example, mutation of | + | A number of mutations completely eliminate calcium uptake by the MCU. For example, mutation of an residue in the <scene name='83/832952/Dxxe_motif/7'>WDXXEP motif</scene>, with the exception of the two "X" residues, altered the highly conserved selectivity filter and completely eliminated calcium uptake.<ref name="Baradaran"/><ref name="Fan"/> Even substituting Glu336 with an aspartate residue significantly changes the dimensions of the pore and inhibits uptake of calcium. Mutation of the secondary tyrosine ring substantially impaired calcium intake and proper protein folding.<ref name="Fan"/> Additional mutations outside the selectivity filter also impacted calcium uptake, including Trp317 (analogous to <scene name='83/832952/New_ones/7'>Trp210</scene> in ''C. europaea'') which has a side chain constituting a primary contact point between TM1 and TM2.<ref name="Fan"/> Mutation of human MCU Phe326 (analogous to <scene name='83/832952/New_ones/8'>Phe218</scene> in ''C. europaea'') or Gly331 of the TM1-TM2 linker (<scene name='83/832952/New_ones/9'>Gly223</scene> in ''C. europaea'') also affected the linker conformation and configuration of the pore entrance and impaired calcium intake.<ref name="Fan"/> |
==Regulation and Inhibition== | ==Regulation and Inhibition== | ||
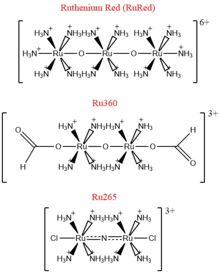
[[Image:Ruthenium_Inhibitors.jpg|220 px|right|thumb|Figure 4: Structures of the ruthenium-based inhibitors of the MCU. Created using ChemDraw Professional 16.0]] | [[Image:Ruthenium_Inhibitors.jpg|220 px|right|thumb|Figure 4: Structures of the ruthenium-based inhibitors of the MCU. Created using ChemDraw Professional 16.0]] | ||
| - | The most well-known and commonly used inhibitor of the MCU is [https://en.wikipedia.org/wiki/Ruthenium_red ruthenium red] (RuRed).<ref name="Woods"/> RuRed is a trinuclear, oxo-bridged complex that effectively inhibits calcium uptake without affecting mitochondrial respiration or calcium efflux. The disadvantage of ruthenium red is its challenging purification.<ref name="Woods"/> Interestingly, a compound identified in an impure version of RuRed, termed [https://en.wikipedia.org/wiki/Ru360 Ru360], was found to be the actual active component as an inhibitor of the MCU (Figure 4). Ru360 is a binuclear, oxo-bridged complex with a similar structure to that of RuRed. | + | The most well-known and commonly used inhibitor of the MCU is [https://en.wikipedia.org/wiki/Ruthenium_red ruthenium red] (RuRed).<ref name="Woods"/> RuRed is a trinuclear, oxo-bridged complex that effectively inhibits calcium uptake without affecting mitochondrial respiration or calcium efflux. The disadvantage of ruthenium red is its challenging purification.<ref name="Woods"/> Interestingly, a compound identified in an impure version of RuRed, termed [https://en.wikipedia.org/wiki/Ru360 Ru360], was found to be the actual active component as an inhibitor of the MCU (Figure 4). Ru360 is a binuclear, oxo-bridged complex with a similar structure to that of RuRed. To increase the cell permeability of Ru360, the derivative Ru265 was subsequently which had twice the cell permeability of Ru360. Ru265 possesses two bridged Ru centers bridged by a nitride ligand (Figure 4).<ref name="Woods"/> |
Recent experiments suggest that Ru360 inhibits calcium uptake through interactions with the <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> motif. However, not much is known about the mode of inhibition. Also, mutations of Asp261 and Ser259 in human MCU were shown to maintain calcium uptake into the matrix, but reduce the inhibitory effect of Ru360, but not Ru265. However, a mutation in a cysteine residue in the <scene name='83/832952/New_ones/4'>NTD</scene> had the opposite effect as it reduced the inhibitory effects of Ru265, but not Ru360 (Figure 4).<ref name="Woods"/> | Recent experiments suggest that Ru360 inhibits calcium uptake through interactions with the <scene name='83/832952/Dxxe_motif/7'>WDXXEP</scene> motif. However, not much is known about the mode of inhibition. Also, mutations of Asp261 and Ser259 in human MCU were shown to maintain calcium uptake into the matrix, but reduce the inhibitory effect of Ru360, but not Ru265. However, a mutation in a cysteine residue in the <scene name='83/832952/New_ones/4'>NTD</scene> had the opposite effect as it reduced the inhibitory effects of Ru265, but not Ru360 (Figure 4).<ref name="Woods"/> | ||
==Medical Relevance== | ==Medical Relevance== | ||
| - | The MCU | + | The MCU is connected with various diseases due to its effect on apoptosis and cell signaling. The overload of the mitochondrial matrix with calcium leads to release of [https://en.wikipedia.org/wiki/Cytochrome_c cytochrome c], overproduction of [https://en.wikipedia.org/wiki/Reactive_oxygen_species reactive oxygen species], mitochondrial swelling, and the opening of the mitochondrial permeability transition pore ([https://en.wikipedia.org/wiki/Mitochondrial_permeability_transition_pore mPTP]) which all lead to apoptotic cell death.<ref name="Woods"/> This connection between mitochondrial calcium and apoptosis makes MCU dysregulation a large contributor to cell death and disease. Calcium machinery in the mitochondria are targets for [https://en.wikipedia.org/wiki/Oncogene proto-oncogenes] and [https://en.wikipedia.org/wiki/Tumor_suppressor tumor suppressors] for this very reason.<ref name="Giorgi"/> Apoptosis can either be induced or repressed. Furthermore, external stimuli can activate receptors in the endoplasmic reticulum that release calcium and activate signal transductions.<ref name="Wang"/> Sequestration of calcium in the mitochondria is vital to shut down these activations, so any impact in movement of calcium ions can cause a wide variety of diseases.<ref name="Wang"/> |
===Neurodegenerative Disorders=== | ===Neurodegenerative Disorders=== | ||
| - | Disruption in calcium homeostasis leads to a wide range of [https://en.wikipedia.org/wiki/Neurodegeneration neurodegenerative disorders]. The MCU complex | + | Disruption in calcium homeostasis leads to a wide range of [https://en.wikipedia.org/wiki/Neurodegeneration neurodegenerative disorders]. The MCU complex plays a role in [https://en.wikipedia.org/wiki/Neuromuscular_disease neuromuscular disease] because of a loss of function of the MICU1 subunit.<ref name="Woods"/> Mutation of MICU1 causes [https://en.wikipedia.org/wiki/Myopathy myopathy], learning difficulties, and progressive movement disorders which can be lethal. In [https://en.wikipedia.org/wiki/Alzheimer%27s_disease Alzheimer's disease], the buildup of [https://en.wikipedia.org/wiki/Amyloid_beta amyloid-β] plaques in the brain leads to increased calcium uptake in [https://en.wikipedia.org/wiki/Neuron neurons] and cell death. Similarly, in early onset [https://en.wikipedia.org/wiki/Parkinson%27s_disease Parkinson's disease], degradation of MICU1 by the ligase [http://proteopedia.org/wiki/index.php/Parkin Parkin] leads to increased mitochondrial calcium uptake, overload, and death. Finally, disrupted glutamate homeostasis in [https://en.wikipedia.org/wiki/Astrocyte astrocytes] and neurons leads to calcium overload and cell death via [https://en.wikipedia.org/wiki/Excitotoxicity excitotoxicity] in Amyotrophic Lateral Sclerosis ([https://en.wikipedia.org/wiki/Amyotrophic_lateral_sclerosis ALS]).<ref name="Woods"/> |
===Diabetes=== | ===Diabetes=== | ||
| - | Calcium homeostasis misregulation | + | Calcium homeostasis misregulation is also instrumental in [https://en.wikipedia.org/wiki/Obesity obesity], [https://en.wikipedia.org/wiki/Insulin_resistance insulin resistance], and [https://en.wikipedia.org/wiki/Type_2_diabetes type-II diabetes].<ref name="Wang"/> The intracellular calcium concentrations in primary [https://en.wikipedia.org/wiki/Adipocyte adipocytes] from obese human subjects are elevated. Any inhibition of downstream calcium signaling could decrease movement of the [http://proteopedia.org/wiki/index.php/GLUT4 GLUT4] glucose transporter and glucose uptake. Additionally, removal of MCU in [https://en.wikipedia.org/wiki/Beta_cell β-cells] in the [https://en.wikipedia.org/wiki/Pancreas pancreas] demonstrated a decrease in cellular ATP concentration following glucose stimulation which resulted in decreased glucose-stimulated insulin secretion. |
===Heart Failure=== | ===Heart Failure=== | ||
Revision as of 19:11, 2 December 2020
Mitochondrial Calcium Uniporter
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 1.22 Baradaran R, Wang C, Siliciano AF, Long SB. Cryo-EM structures of fungal and metazoan mitochondrial calcium uniporters. Nature. 2018 Jul 11. pii: 10.1038/s41586-018-0331-8. doi:, 10.1038/s41586-018-0331-8. PMID:29995857 doi:http://dx.doi.org/10.1038/s41586-018-0331-8
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 2.9 Woods JJ, Wilson JJ. Inhibitors of the mitochondrial calcium uniporter for the treatment of disease. Curr Opin Chem Biol. 2019 Dec 20;55:9-18. doi: 10.1016/j.cbpa.2019.11.006. PMID:31869674 doi:http://dx.doi.org/10.1016/j.cbpa.2019.11.006
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Giorgi C, Marchi S, Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol. 2018 Nov;19(11):713-730. doi: 10.1038/s41580-018-0052-8. PMID:30143745 doi:http://dx.doi.org/10.1038/s41580-018-0052-8
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 Wang CH, Wei YH. Role of mitochondrial dysfunction and dysregulation of Ca(2+) homeostasis in the pathophysiology of insulin resistance and type 2 diabetes. J Biomed Sci. 2017 Sep 7;24(1):70. doi: 10.1186/s12929-017-0375-3. PMID:28882140 doi:http://dx.doi.org/10.1186/s12929-017-0375-3
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 Fan C, Fan M, Orlando BJ, Fastman NM, Zhang J, Xu Y, Chambers MG, Xu X, Perry K, Liao M, Feng L. X-ray and cryo-EM structures of the mitochondrial calcium uniporter. Nature. 2018 Jul 11. pii: 10.1038/s41586-018-0330-9. doi:, 10.1038/s41586-018-0330-9. PMID:29995856 doi:http://dx.doi.org/10.1038/s41586-018-0330-9
Student Contributors
Ryan Heumann
Lizzy Ratz
Holly Rowe
Madi Summers
Rieser Wells