This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
1db1
From Proteopedia
| Line 1: | Line 1: | ||
[[Image:1db1.gif|left|200px]] | [[Image:1db1.gif|left|200px]] | ||
| - | + | <!-- | |
| - | + | The line below this paragraph, containing "STRUCTURE_1db1", creates the "Structure Box" on the page. | |
| - | + | You may change the PDB parameter (which sets the PDB file loaded into the applet) | |
| - | + | or the SCENE parameter (which sets the initial scene displayed when the page is loaded), | |
| - | | | + | or leave the SCENE parameter empty for the default display. |
| - | | | + | --> |
| - | + | {{STRUCTURE_1db1| PDB=1db1 | SCENE= }} | |
| - | + | ||
| - | + | ||
| - | }} | + | |
'''CRYSTAL STRUCTURE OF THE NUCLEAR RECEPTOR FOR VITAMIN D COMPLEXED TO VITAMIN D''' | '''CRYSTAL STRUCTURE OF THE NUCLEAR RECEPTOR FOR VITAMIN D COMPLEXED TO VITAMIN D''' | ||
| Line 30: | Line 27: | ||
[[Category: Rochel, N.]] | [[Category: Rochel, N.]] | ||
[[Category: Wurtz, J M.]] | [[Category: Wurtz, J M.]] | ||
| - | [[Category: | + | [[Category: Complex]] |
| - | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Fri May 2 13:38:50 2008'' | |
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | |
Revision as of 10:38, 2 May 2008
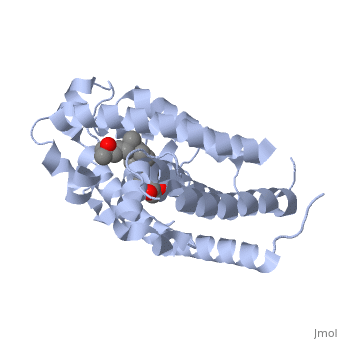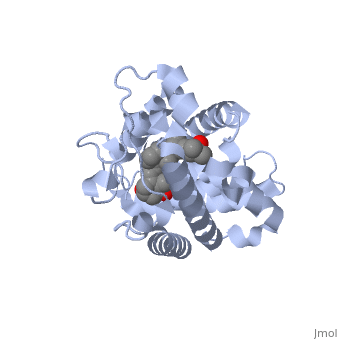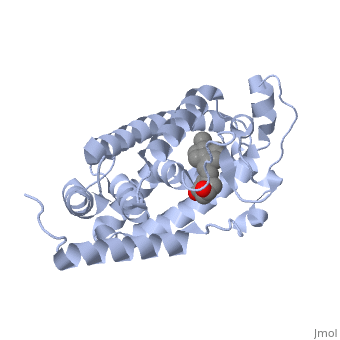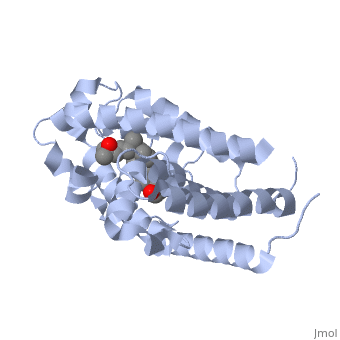
CRYSTAL STRUCTURE OF THE NUCLEAR RECEPTOR FOR VITAMIN D COMPLEXED TO VITAMIN D
Overview
The action of 1 alpha, 25-dihydroxyvitamin D3 is mediated by its nuclear receptor (VDR), a ligand-dependent transcription regulator. We report the 1.8 A resolution crystal structure of the complex between a VDR ligand-binding domain (LBD) construct lacking the highly variable VDR-specific insertion domain and vitamin D. The construct exhibits the same binding affinity for vitamin D and transactivation ability as the wild-type protein, showing that the N-terminal part of the LBD is essential for its structural and functional integrity while the large insertion peptide is dispensable. The structure reveals the active conformation of the bound ligand and allows understanding of the different binding properties of some synthetic analogs.
About this Structure
1DB1 is a Single protein structure of sequence from Homo sapiens. Full crystallographic information is available from OCA.
Reference
The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand., Rochel N, Wurtz JM, Mitschler A, Klaholz B, Moras D, Mol Cell. 2000 Jan;5(1):173-9. PMID:10678179 Page seeded by OCA on Fri May 2 13:38:50 2008