Sandbox Reserved 1649
From Proteopedia
| Line 17: | Line 17: | ||
'''Ligand binding domain (LBD)''' | '''Ligand binding domain (LBD)''' | ||
| - | LBD is constituted of two domains S1 (located juste upstream M1 transmembrane domain) and S2 and has affinity for glutamate or sometime glycine. Positive charge of amino-group of the agonist bind to negative charges residue of the pocket D731. In [https://proteopedia.org/wiki/index.php/Glutamate_receptor_%28GluA2%29 GlurR], negative charge amino acid is a E731 and is able to form salt bridge with agonist. In NR2A D731 (which corresponds to <scene name='86/868182/D213/1'>D213</scene>) is not able to do salt bridge with amino group because aspartate is one methylene lacking to do it. Amino group of agonist is stabilized by water mediated hydrogen bonds to amino acid Y761 (which corresponds to <scene name='86/868182/Y243/1'>Y243</scene>) and E413 (which correspond to <scene name='86/868182/E14/1'>E14</scene>).<scene name='86/868182/Y243_et_e14/1'>Click here if you want to see E14 and Y243 together</scene>. The high affinity for glutamate agonist may be because of van der Walls contact between γ-carboxylate group of glutamate and Y730 of S2 domain which is conserved in NR2 protein.<ref name="LBD">DOI 10.1038/nature04089</ref> Amino-group of glutamate also interacts with <scene name='86/868182/T114/1'>T114</scene> and <scene name='86/868182/S112/1'>S112</scene>. | + | LBD is constituted of two domains S1 (located juste upstream M1 transmembrane domain) and S2 and has affinity for glutamate or sometime glycine. Positive charge of amino-group of the agonist bind to negative charges residue of the pocket D731. In [https://proteopedia.org/wiki/index.php/Glutamate_receptor_%28GluA2%29 GlurR], negative charge amino acid is a E731 and is able to form salt bridge with agonist. In NR2A D731 (which corresponds to <scene name='86/868182/D213/1'>D213</scene>) is not able to do salt bridge with amino group because aspartate is one methylene lacking to do it. Amino group of agonist is stabilized by water mediated hydrogen bonds to amino acid Y761 (which corresponds to <scene name='86/868182/Y243/1'>Y243</scene>) and E413 (which correspond to <scene name='86/868182/E14/1'>E14</scene>).<scene name='86/868182/Y243_et_e14/1'>Click here if you want to see E14 and Y243 together</scene>. The high affinity for glutamate agonist may be because of van der Walls contact between γ-carboxylate group of glutamate and Y730 of S2 domain which is conserved in NR2 protein.<ref name="LBD">DOI 10.1038/nature04089</ref> Amino-group of glutamate also interacts with <scene name='86/868182/T114/1'>T114</scene> and <scene name='86/868182/S112/1'>S112</scene>. <scene name='86/868182/T114_et_s112/1'>(T114 and S112 together)</scene> |
On the other hand, <scene name='86/868182/Aa_in_interaction_with_nr1/1'>amino acids</scene> from this domain interact with NR1 (see NR1/NR2A complex part). | On the other hand, <scene name='86/868182/Aa_in_interaction_with_nr1/1'>amino acids</scene> from this domain interact with NR1 (see NR1/NR2A complex part). | ||
Revision as of 14:11, 15 January 2021
|
Contents |
NR2A (2A5S)
NR2A is a protein which is a part of NMDA receptors, heterodimer channels composed of four subunits. Indeed, NMDA receptors are made of the association between two NR2 and two NR1 proteins. NMDA receptors play a key role in mammalian central nervous system, as they act in Ca2+ influx in synapses in response to glutamate and glycine binding. Their role is essential for learning and memory. Variety of NR2 allows modulation of NMDAr. In the other hand, NMDA receptor is related to AMPA receptor in the same synapse.
Structure and Function
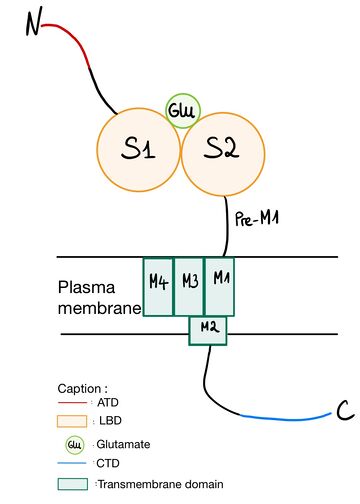
NR2A (GluN2A) is composed of Amino-terminal domain (ATD), segments S1 and S2 which formed ligand binding domain of glutamate, three transmembrane helices (M1, M3,and M4), a cytoplasmic re-entrant pore loop (M2), and an intracellular C-terminal domain (CTD).Amino-terminal domain (ATD)
The ATD is constituted by first 383 amino acids of NR2A. ATD is an alpha and beta protein class. Structure is bilobed and form clam-shell like structure which consists in two lobes linked by a flexible hinge region defining a central groove. [1] Zn2+ may insert between 2 lobes and induces closure of channel by changing conformation of ATD. Zn increases affinity of glutamate on the LBD which reminds the desensitization of AMPA and Kainate receptor. [2] [3] ATD allows to modulate NMDA receptor. Difference between different NR2 is mainly regulated by ATD, because diversity of ATD can modulate traffic in endoplasmic reticulum and then affect the localization of NMDAr. ATD of NR2A increases glutamate affinity, control channel’s opening with high probability and open duration, control glutamate deactivation time course.[4]
Ligand binding domain (LBD)
LBD is constituted of two domains S1 (located juste upstream M1 transmembrane domain) and S2 and has affinity for glutamate or sometime glycine. Positive charge of amino-group of the agonist bind to negative charges residue of the pocket D731. In GlurR, negative charge amino acid is a E731 and is able to form salt bridge with agonist. In NR2A D731 (which corresponds to ) is not able to do salt bridge with amino group because aspartate is one methylene lacking to do it. Amino group of agonist is stabilized by water mediated hydrogen bonds to amino acid Y761 (which corresponds to ) and E413 (which correspond to ).. The high affinity for glutamate agonist may be because of van der Walls contact between γ-carboxylate group of glutamate and Y730 of S2 domain which is conserved in NR2 protein.[5] Amino-group of glutamate also interacts with and .
On the other hand, from this domain interact with NR1 (see NR1/NR2A complex part).
Transmembrane domain The transmembrane domain is organized into 4 parts (from M1 to M4). M1 connects the N terminal domain to M2. M2 forms a reentrant loop contributing to the pore. The S1 segment of the N terminal domain intertwines with the S2 segment of the GlnBP-type domain in the extracellular loop M3 - M4 to form the glutamate binding pocket. On the other hand, desensitization of NMDA receptors is affected by residues near or inside the binding pocket as well as by residues in M2 that line the pore and the M3 loop - M4 is not responsible for the specificity of the NR2 subunit of glycine independent desensitization.[6] M2 loop is a channel-lining loop and located in transmembrane domain. Two asparagines are located on N site of the domain and block Mg2+ and are permeable of Ca2+ [7] Ethanol acts as an inhibitor on NMDAr. Phenylalanine at position 639 in the M3 part of the transmembrane domain of NR2A interacts with the latter.[8]
CTD
CTD domain is the less conserved of NR2 domain and like ATD allows différents localisation of NMDAr thanks to reticulum endoplasmic trafficking. It is indispensable to receptor surface dynamic and activation of specific signaling. CTD phosphorylation can modulate NMDAr, for instance it is useful for endocytosis during glutamate binding on LBD. [7]
Regulation
NR1/NR2A complex
NR2A can be found in some NMDA receptors, in which NR2A is associated to NR1. NMDA receptors are necessarily heteromers of 4 subunits, organized as a dimer of dimers. These dimers are heterodimers constituted with glycine binding NR1 and glutamate binding subunits NR2.
NR1 and NR2A are assembled in a dimer, arranged in a back-to-back fashion, thanks to interactions between three different domains on each subunit: sites I, II and III. - Site II: The link between NR2A and NR1 is made by at least three amino acids: E530 () makes a salt bridge with R755 of NR1, F524 () binds K531 of NR1 by a hydrogen bond on the backbone carbonyl oxygen, and P257. - Sites I and III: the binding is established by hydrophobic residues (I514 (), V526 (), L777, L780 present on helices D and J), or by polar contacts
Depending on the kind of NR2 (A-D) linked to NR1, the affinity of NR1 for glycine can be affected. Moreover, for a particular combination of NR1 and NR2 subunits, a negative cooperativity has been observed between glycine and glutamate binding. This leads to consider a possible allosteric coupling between NR1 and NR2. Thus, the importance of the structure of NR2A to make contacts with NR1 is obvious. But mechanistic explanation about the role that subunit-subunit contacts might have in NMDA receptor activity has not been found yet.
Since the opening of the NMDA channel requires both glutamate and glycine, respectively detected by NR2 and NR1, the association between NR1 and NR2A is primordial.
Mutations
The C-terminal truncation of NR2 subunits in NMDA receptors is an interesting mutation. Indeed, the C-terminal truncation of NR2A influences the functioning of the NMDA receptor. Several experiments were carried out and made it possible to conclude on this influence. First, mice with truncated NR2A subunits have been shown to still have a functional receptor channel, but NR2A ΔC / ΔC mice appear to be altered in the cellular signal transduction events involved in the induction of LTP (potentiation long-term). Despite the presence of the full-length NR2B subunit, the C-terminal truncation of the NR2A subunit altered the signal transduction mediated by NMDAR. Second, the C-terminal truncation of NR2A has been shown to alter fear in a particular setting. Indeed, the experiment was to put mutant and wild mice through stepwise avoidance training and put them in water. Thus, the researchers concluded that mutant mice have a significantly reduced latency time to leave the safe platform compared to wild mice and that mutant mice exhibit water balance deficits.
The C-terminal truncation of NR2A therefore has consequences on synaptic plasticity and synaptic reorganization during the recording of hippocampal LTP, on the conditioning of fear and also on motor coordination. [9]
NMDA receptors are inhibited by ethanol. Mutagenesis of an F residue in the transmembrane domain has made it possible to characterize the role of the latter in this inhibition. Structurally, there is a small loop of 150 amino acids between M3 and M4. On the other hand, the phenylalanine residue at position 639 is replaced by an alanine in the transmembrane domain (in part M3) and the results show significantly less inhibition by ethanol of NMDA compared to wild type receptors.[8]
Disease
Anti-nuclear antibodies produced in systemic or systemic lupus erythematosus (SLE, SLE) interact with the NR2A subunit of the NMDA receptor. This is because the Asp / Glu-Trp-Asp / Glu-Tyr-Ser / Gly pentapeptide is a molecular mimic of double-stranded DNA, so antibodies produced in a patient with SLE recognize this pentapeptide. The latter is also present in the structure of NR2A. Thus, these antibodies cross-interact with NR2A and therefore the NMDA receptor. This interaction can signal neuronal death by an excitotoxic mechanism. More generally, NMDA receptor dysfunction is implicated in multiple brain disorders, such as stroke, chronic pain, and schizophrenia. [10]
</StructureSection>
References
- ↑ Zhu S, Stroebel D, Yao CA, Taly A, Paoletti P. Allosteric signaling and dynamics of the clamshell-like NMDA receptor GluN1 N-terminal domain. Nat Struct Mol Biol. 2013 Apr;20(4):477-85. doi: 10.1038/nsmb.2522. Epub 2013 Mar, 3. PMID:23454977 doi:http://dx.doi.org/10.1038/nsmb.2522
- ↑ Paoletti P, Perin-Dureau F, Fayyazuddin A, Le Goff A, Callebaut I, Neyton J. Molecular organization of a zinc binding n-terminal modulatory domain in a NMDA receptor subunit. Neuron. 2000 Dec;28(3):911-25. doi: 10.1016/s0896-6273(00)00163-x. PMID:11163276 doi:http://dx.doi.org/10.1016/s0896-6273(00)00163-x
- ↑ Gielen M. [Molecular operation of ionotropic glutamate receptors: proteins that mediate the excitatory synaptic neurotransmission]. Med Sci (Paris). 2010 Jan;26(1):65-72. doi: 10.1051/medsci/201026165. PMID:20132777 doi:http://dx.doi.org/10.1051/medsci/201026165
- ↑ Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci. 2009 Sep 30;29(39):12045-58. doi: 10.1523/JNEUROSCI.1365-09.2009. PMID:19793963 doi:http://dx.doi.org/10.1523/JNEUROSCI.1365-09.2009
- ↑ Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005 Nov 10;438(7065):185-92. PMID:16281028 doi:10.1038/nature04089
- ↑ Krupp JJ, Vissel B, Heinemann SF, Westbrook GL. N-terminal domains in the NR2 subunit control desensitization of NMDA receptors. Neuron. 1998 Feb;20(2):317-27. doi: 10.1016/s0896-6273(00)80459-6. PMID:9491992 doi:http://dx.doi.org/10.1016/s0896-6273(00)80459-6
- ↑ 7.0 7.1 Franchini L, Carrano N, Di Luca M, Gardoni F. Synaptic GluN2A-Containing NMDA Receptors: From Physiology to Pathological Synaptic Plasticity. Int J Mol Sci. 2020 Feb 24;21(4). pii: ijms21041538. doi: 10.3390/ijms21041538. PMID:32102377 doi:http://dx.doi.org/10.3390/ijms21041538
- ↑ 8.0 8.1 Ronald KM, Mirshahi T, Woodward JJ. Ethanol inhibition of N-methyl-D-aspartate receptors is reduced by site-directed mutagenesis of a transmembrane domain phenylalanine residue. J Biol Chem. 2001 Nov 30;276(48):44729-35. doi: 10.1074/jbc.M102800200. Epub 2001, Sep 25. PMID:11572853 doi:http://dx.doi.org/10.1074/jbc.M102800200
- ↑ Sprengel R, Suchanek B, Amico C, Brusa R, Burnashev N, Rozov A, Hvalby O, Jensen V, Paulsen O, Andersen P, Kim JJ, Thompson RF, Sun W, Webster LC, Grant SG, Eilers J, Konnerth A, Li J, McNamara JO, Seeburg PH. Importance of the intracellular domain of NR2 subunits for NMDA receptor function in vivo. Cell. 1998 Jan 23;92(2):279-89. doi: 10.1016/s0092-8674(00)80921-6. PMID:9458051 doi:http://dx.doi.org/10.1016/s0092-8674(00)80921-6
- ↑ DeGiorgio LA, Konstantinov KN, Lee SC, Hardin JA, Volpe BT, Diamond B. A subset of lupus anti-DNA antibodies cross-reacts with the NR2 glutamate receptor in systemic lupus erythematosus. Nat Med. 2001 Nov;7(11):1189-93. doi: 10.1038/nm1101-1189. PMID:11689882 doi:http://dx.doi.org/10.1038/nm1101-1189