This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
PROTAC
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| + | <StructureSection load='' size='340' side='right' caption='' scene=''> | ||
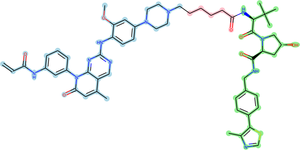
A Proteolysis-Targeting Chimera (PROTAC) is a small protein with two active domains joined by a linker. PROTACs represent an alternative to conventional enzyme inhibitors, by enabling [[Proteasome|proteasome]] induced degradation of the target protein <ref>pmid 28288108</ref>. | A Proteolysis-Targeting Chimera (PROTAC) is a small protein with two active domains joined by a linker. PROTACs represent an alternative to conventional enzyme inhibitors, by enabling [[Proteasome|proteasome]] induced degradation of the target protein <ref>pmid 28288108</ref>. | ||
| Line 8: | Line 9: | ||
==PROTACpedia== | ==PROTACpedia== | ||
PROTACpedia <ref>PROTACpedia https://protacpedia.weizmann.ac.il/, a resource of manually curated data on Proteolysis Targeting Chimeras (PROTACs).</ref> is a collaborative and comprehensive, high-quality and freely accessible resource of manually curated data on Proteolysis Targeting Chimeras (PROTACs). The search can be performed by various parameters such as Target name, UniProt ID, SMILES substructure or by various details regarding a publication. | PROTACpedia <ref>PROTACpedia https://protacpedia.weizmann.ac.il/, a resource of manually curated data on Proteolysis Targeting Chimeras (PROTACs).</ref> is a collaborative and comprehensive, high-quality and freely accessible resource of manually curated data on Proteolysis Targeting Chimeras (PROTACs). The search can be performed by various parameters such as Target name, UniProt ID, SMILES substructure or by various details regarding a publication. | ||
| - | + | </StructureSection> | |
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 10:36, 28 February 2021
| |||||||||||
References
- ↑ Gadd MS, Testa A, Lucas X, Chan KH, Chen W, Lamont DJ, Zengerle M, Ciulli A. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol. 2017 Mar 13. doi: 10.1038/nchembio.2329. PMID:28288108 doi:http://dx.doi.org/10.1038/nchembio.2329
- ↑ PRosettaC https://prosettac.weizmann.ac.il, a computational resource for the prediction of PROTAC-induced ternary complexes.
- ↑ Zaidman D, Prilusky J, London N. PRosettaC: Rosetta based modeling of PROTAC mediated ternary complexes. J Chem Inf Model. 2020 Sep 25. doi: 10.1021/acs.jcim.0c00589. PMID:32976709 doi:http://dx.doi.org/10.1021/acs.jcim.0c00589
- ↑ PROTACpedia https://protacpedia.weizmann.ac.il/, a resource of manually curated data on Proteolysis Targeting Chimeras (PROTACs).