PROTAC
From Proteopedia
(Difference between revisions)
| Line 27: | Line 27: | ||
==PROTACpedia== | ==PROTACpedia== | ||
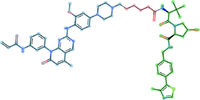
| - | [[Image:PROTAC_70.png|thumb|right| | + | [[Image:PROTAC_70.png|thumb|right|200px|Representative PROTAC from [https://protacpedia.weizmann.ac.il/ptcb/detp?i=70 PROTACpedia] targeting EGFR Epidermal growth factor receptor P00533. <span class="bg-lightblue"><b>target binding ligand</b></span><span class="bg-lightmagenta"><b>linker</b></span><span class="bg-lightgreen"><b>E3 binder</b></span>]] |
PROTACpedia <ref>PROTACpedia https://protacpedia.weizmann.ac.il/, a resource of manually curated data on Proteolysis Targeting Chimeras (PROTACs).</ref> is a collaborative and comprehensive, high-quality and freely accessible resource of manually curated data on Proteolysis Targeting Chimeras (PROTACs). The search can be performed by various parameters such as Target name, UniProt ID, SMILES substructure or by various details regarding a publication. | PROTACpedia <ref>PROTACpedia https://protacpedia.weizmann.ac.il/, a resource of manually curated data on Proteolysis Targeting Chimeras (PROTACs).</ref> is a collaborative and comprehensive, high-quality and freely accessible resource of manually curated data on Proteolysis Targeting Chimeras (PROTACs). The search can be performed by various parameters such as Target name, UniProt ID, SMILES substructure or by various details regarding a publication. | ||
Revision as of 18:55, 28 February 2021
| |||||||||||
References
- ↑ Gadd MS, Testa A, Lucas X, Chan KH, Chen W, Lamont DJ, Zengerle M, Ciulli A. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol. 2017 Mar 13. doi: 10.1038/nchembio.2329. PMID:28288108 doi:http://dx.doi.org/10.1038/nchembio.2329
- ↑ PRosettaC https://prosettac.weizmann.ac.il, a computational resource for the prediction of PROTAC-induced ternary complexes.
- ↑ Zaidman D, Prilusky J, London N. PRosettaC: Rosetta based modeling of PROTAC mediated ternary complexes. J Chem Inf Model. 2020 Sep 25. doi: 10.1021/acs.jcim.0c00589. PMID:32976709 doi:http://dx.doi.org/10.1021/acs.jcim.0c00589
- ↑ Drummond ML, Henry A, Li H, Williams CI. Improved Accuracy for Modeling PROTAC-Mediated Ternary Complex Formation and Targeted Protein Degradation via New In Silico Methodologies. J Chem Inf Model. 2020 Oct 26;60(10):5234-5254. doi: 10.1021/acs.jcim.0c00897., Epub 2020 Oct 12. PMID:32969649 doi:http://dx.doi.org/10.1021/acs.jcim.0c00897
- ↑ Bai N, Miller SA, Andrianov GV, Yates M, Kirubakaran P, Karanicolas J. Rationalizing PROTAC-Mediated Ternary Complex Formation Using Rosetta. J Chem Inf Model. 2021 Feb 24. doi: 10.1021/acs.jcim.0c01451. PMID:33625214 doi:http://dx.doi.org/10.1021/acs.jcim.0c01451
- ↑ PROTACpedia https://protacpedia.weizmann.ac.il/, a resource of manually curated data on Proteolysis Targeting Chimeras (PROTACs).