User:Madison Unger/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 22: | Line 22: | ||
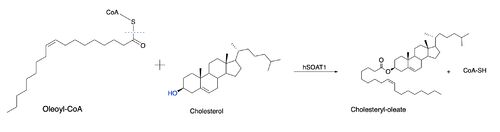
ACATs are enzymes that can be allosterically activated by sterol molecules like cholesterol. There are two proposed binding sites for cholesterol, with each binding site being distinctively different. One site is the substrate active site mentioned above and the other site is the allosteric binding site. The allosteric binding site has the ability to direct feedback regulation over the concentration of cholesterol in the endoplasmic reticulum. | ACATs are enzymes that can be allosterically activated by sterol molecules like cholesterol. There are two proposed binding sites for cholesterol, with each binding site being distinctively different. One site is the substrate active site mentioned above and the other site is the allosteric binding site. The allosteric binding site has the ability to direct feedback regulation over the concentration of cholesterol in the endoplasmic reticulum. | ||
| - | In order to act as an allosteric activator, the sterol molecules must contain a highly conserved 3-beta hydroxyl group at the first steroid ring (ring A) <ref>PMID:25218443</ref> . An activator must also contain a conserved iso-octyl side chain to efficiently activate ACAT. The conserved 3-beta hydroxyl group allows for binding of the cholesterol to cause conformational changes to the ACAT dimer. Upon conformational changes, the rate of esterification is increased. | + | In order to act as an allosteric activator, the [http://en.wikipedia.org/wiki/Sterol sterol] molecules must contain a highly conserved 3-beta hydroxyl group at the first steroid ring (ring A) <ref>PMID:25218443</ref> . An activator must also contain a conserved iso-octyl side chain to efficiently activate ACAT. The conserved 3-beta hydroxyl group allows for binding of the cholesterol to cause conformational changes to the ACAT dimer. Upon conformational changes, the rate of esterification is increased. |
===Tunnels=== | ===Tunnels=== | ||
Revision as of 19:56, 29 March 2021
ACAT/SOAT
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
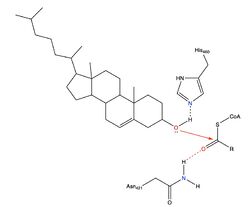
- ↑ Guan C, Niu Y, Chen SC, Kang Y, Wu JX, Nishi K, Chang CCY, Chang TY, Luo T, Chen L. Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nat Commun. 2020 May 18;11(1):2478. doi: 10.1038/s41467-020-16288-4. PMID:32424158 doi:http://dx.doi.org/10.1038/s41467-020-16288-4
- ↑ Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ Rogers MA, Liu J, Song BL, Li BL, Chang CC, Chang TY. Acyl-CoA:cholesterol acyltransferases (ACATs/SOATs): Enzymes with multiple sterols as substrates and as activators. J Steroid Biochem Mol Biol. 2015 Jul;151:102-7. doi: 10.1016/j.jsbmb.2014.09.008., Epub 2014 Sep 12. PMID:25218443 doi:http://dx.doi.org/10.1016/j.jsbmb.2014.09.008
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
Student Contributors
- Leah Goehring
- Gabby Smith
- Anna Campbell