User:Madison Unger/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 25: | Line 25: | ||
===Tunnels=== | ===Tunnels=== | ||
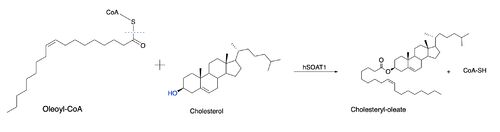
| - | The nine transmembrane segments create a cytosolic tunnel and a transmembrane tunnel that meet at the site of catalysis (active site). Acyl-coenzyme A enters the active site though the cytosolic tunnel and cholesterol enters through the transmembrane tunnel. These then meet in the catalytic site to react and form the cholesteryl ester. When exiting the catalytic site, the free CoA is able to release through the cytosolic tunnel to the cytosol and the cholesterol ester is able to release through the transmembrane tunnel to the membrane or through the lumen tunnel to the lumen. Certain residues that line the transmembrane tunnel are important in ACAT activity (E259, E263, R262, P304, L306, V423, V424, M265, I261, and H460). | + | The nine transmembrane segments create a cytosolic tunnel and a transmembrane tunnel that meet at the site of catalysis (active site). Acyl-coenzyme A enters the active site though the cytosolic tunnel and cholesterol enters through the transmembrane tunnel. These then meet in the catalytic site to react and form the cholesteryl ester. When exiting the catalytic site, the free CoA is able to release through the cytosolic tunnel to the cytosol and the cholesterol ester is able to release through the <scene name='87/877505/Active_site_residues/2'>transmembrane tunnel</scene> to the membrane or through the lumen tunnel to the lumen. Certain residues that line the transmembrane tunnel are important in ACAT activity (E259, E263, R262, P304, L306, V423, V424, M265, I261, and H460). |
== Disease == | == Disease == | ||
[http://https://proteopedia.org/wiki/index.php/Dipeptidyl_peptidase_IV Other Protopedia Page] | [http://https://proteopedia.org/wiki/index.php/Dipeptidyl_peptidase_IV Other Protopedia Page] | ||
Revision as of 20:06, 29 March 2021
ACAT/SOAT
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Guan C, Niu Y, Chen SC, Kang Y, Wu JX, Nishi K, Chang CCY, Chang TY, Luo T, Chen L. Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nat Commun. 2020 May 18;11(1):2478. doi: 10.1038/s41467-020-16288-4. PMID:32424158 doi:http://dx.doi.org/10.1038/s41467-020-16288-4
- ↑ Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ Rogers MA, Liu J, Song BL, Li BL, Chang CC, Chang TY. Acyl-CoA:cholesterol acyltransferases (ACATs/SOATs): Enzymes with multiple sterols as substrates and as activators. J Steroid Biochem Mol Biol. 2015 Jul;151:102-7. doi: 10.1016/j.jsbmb.2014.09.008., Epub 2014 Sep 12. PMID:25218443 doi:http://dx.doi.org/10.1016/j.jsbmb.2014.09.008
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
Student Contributors
- Leah Goehring
- Gabby Smith
- Anna Campbell