We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Leanne Price/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 20: | Line 20: | ||
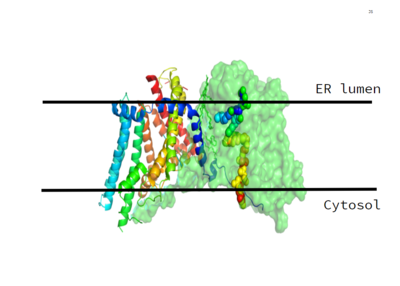
DGAT consists of two domains, one cytoplasmic and one luminal. The cytoplasmic domain interacts with the interior of the cell and relays signals. The luminal domain senses misfolded proteins. The structure of DGAT consists of two protein chains, one ligand, two polymers, eighteen alpha helices and zero beta sheets. The transmembrane helices form a large central cavity within the membrane that opens to the bilayer via a wide lateral gate. Through openings on the cytosolic and luminal sides of DGAT, this central cavity is also accessible. The majority of the transmembrane helices present within the structure also form a concave-shaped ridge on either side of the membrane. These aspects of the domain structure are deemed as the 'MBOAT core'. Within this core, a tunnel-like region, similar to a binding pocket, is also present. Access to the active site of DGAT by substrates is done through the lateral gate, which lies on the ER lumen side, within the membrane. | DGAT consists of two domains, one cytoplasmic and one luminal. The cytoplasmic domain interacts with the interior of the cell and relays signals. The luminal domain senses misfolded proteins. The structure of DGAT consists of two protein chains, one ligand, two polymers, eighteen alpha helices and zero beta sheets. The transmembrane helices form a large central cavity within the membrane that opens to the bilayer via a wide lateral gate. Through openings on the cytosolic and luminal sides of DGAT, this central cavity is also accessible. The majority of the transmembrane helices present within the structure also form a concave-shaped ridge on either side of the membrane. These aspects of the domain structure are deemed as the 'MBOAT core'. Within this core, a tunnel-like region, similar to a binding pocket, is also present. Access to the active site of DGAT by substrates is done through the lateral gate, which lies on the ER lumen side, within the membrane. | ||
| + | |||
| + | [[Image:Image 3.PNG|400 px|left|thumb|Figure 2. Shows the location of the lateral gate as the entrance to the T tunnel via the ER lumen side of the membrane. ]] | ||
| Line 33: | Line 35: | ||
==Mechanism== | ==Mechanism== | ||
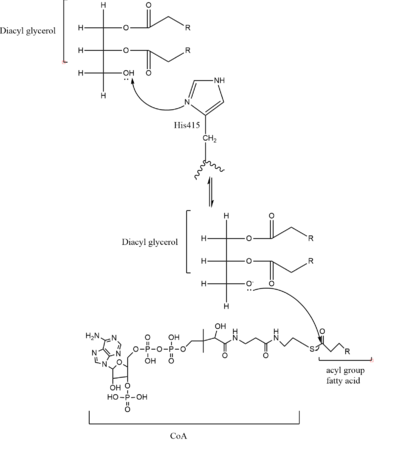
| - | [[Image:Image_2.PNG|400 px|left|thumb|Figure | + | [[Image:Image_2.PNG|400 px|left|thumb|Figure 3. Mechanism for DGAT]] |
In the DGAT mechanism, the diglyceride serves as the nucleophile. While the acyl group of the CoA enzyme serves as the electrophile. The lone pair on the last hydroxyl group present on the glycerol of the diglyceride attacks the thioester bond of the acyl-CoA enzyme. This attack breaks the sulfur-carbon bond, a weak bond that is easily breakable. This allows the acyl group of the acyl-CoA enzyme to attach to the diglyceride, creating a triglyceride. While the CoA group then serves as the leaving group. | In the DGAT mechanism, the diglyceride serves as the nucleophile. While the acyl group of the CoA enzyme serves as the electrophile. The lone pair on the last hydroxyl group present on the glycerol of the diglyceride attacks the thioester bond of the acyl-CoA enzyme. This attack breaks the sulfur-carbon bond, a weak bond that is easily breakable. This allows the acyl group of the acyl-CoA enzyme to attach to the diglyceride, creating a triglyceride. While the CoA group then serves as the leaving group. | ||
Revision as of 19:59, 30 March 2021
DGAT Human
| |||||||||||
References
- ↑ Wang L, Qian H, Nian Y, Han Y, Ren Z, Zhang H, Hu L, Prasad BVV, Laganowsky A, Yan N, Zhou M. Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature. 2020 May;581(7808):329-332. doi: 10.1038/s41586-020-2280-2. Epub 2020 May, 13. PMID:32433610 doi:http://dx.doi.org/10.1038/s41586-020-2280-2
- ↑ Sui X, Wang K, Gluchowski NL, Elliott SD, Liao M, Walther TC, Farese RV Jr. Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature. 2020 May;581(7808):323-328. doi: 10.1038/s41586-020-2289-6. Epub 2020 May, 13. PMID:32433611 doi:http://dx.doi.org/10.1038/s41586-020-2289-6
Student Contributors
- Justin Smith
- Eloi Bigirimana
- Leanne Price