We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Megan Leaman/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 26: | Line 26: | ||
=== Transition State Stabilization === | === Transition State Stabilization === | ||
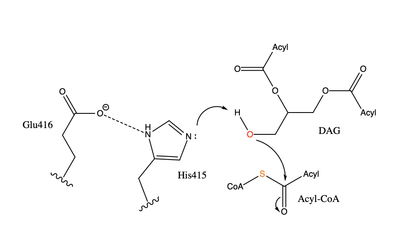
| - | While there is not universal agreement on whether Glu416 or Gln465 provide transition state stabilization for the acyl transfer activity of the enzyme, it is believed that Glu416 is more likely to provide stabilization due to positive findings upon mutations of that residue. Point mutations were made to various residues in the active site including Glu416. Mutations to the entrance of the binding site have a smaller impact on the functionality of the enzyme than mutation to the rest of the active site, while mutations to His415 and Glu416 abolish enzymatic activity completely | + | While there is not universal agreement on whether Glu416 or Gln465 provide transition state stabilization for the acyl transfer activity of the enzyme, it is believed that Glu416 is more likely to provide stabilization due to positive findings upon mutations of that residue. Point mutations were made to various residues in the active site including Glu416. Mutations to the entrance of the binding site have a smaller impact on the functionality of the enzyme than mutation to the rest of the active site, while mutations to His415 and Glu416 abolish enzymatic activity completely. This suggests that His415 and Glu416 are essential to the reaction whereas Gln465 can be changed without significantly impacting the functionality of the enzyme. |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<ref name=”Ransey”>PMID:28504306</ref> | <ref name=”Ransey”>PMID:28504306</ref> | ||
| + | |||
<references/> | <references/> | ||
==Student Contributors== | ==Student Contributors== | ||
Revision as of 21:36, 4 April 2021
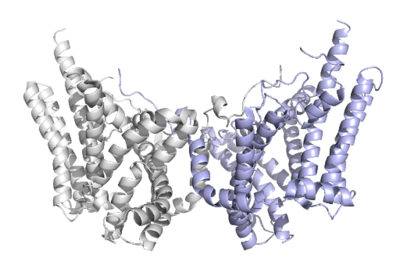
Human Diacylglycerol O-Transferase 1
| |||||||||||
References
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
Student Contributors
- Megan Leaman
- Grace Hall
- Karina Latsko