User:Betsy Johns/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 19: | Line 19: | ||
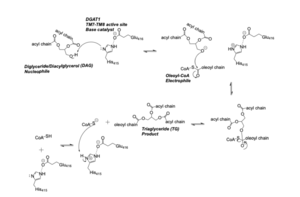
The <scene name='87/877509/Active_site/2'>Active Site Residues</scene> are Asn378, Gln437, Gln465, His415, and Met434. The active site of DGAT serves its catalytic function by placing the His415 residue in close proximity to the acyl-CoA in order to cleave its ester bond and thus bind the fatty acid to the diacylglycerol. The conserved His415 is able to act catalytically due to a charge relay system, where the neighboring Glu416, due to its negative charge pulls electrons on histidine at the N1 position, making the N3 position more nucleophilic. This nitrogen will then deprotonate DAG so it can begin its attack on Acyl-CoA through acyl substitution. The catalytic mechanism for DGAT is shown in Figure 1. | The <scene name='87/877509/Active_site/2'>Active Site Residues</scene> are Asn378, Gln437, Gln465, His415, and Met434. The active site of DGAT serves its catalytic function by placing the His415 residue in close proximity to the acyl-CoA in order to cleave its ester bond and thus bind the fatty acid to the diacylglycerol. The conserved His415 is able to act catalytically due to a charge relay system, where the neighboring Glu416, due to its negative charge pulls electrons on histidine at the N1 position, making the N3 position more nucleophilic. This nitrogen will then deprotonate DAG so it can begin its attack on Acyl-CoA through acyl substitution. The catalytic mechanism for DGAT is shown in Figure 1. | ||
| - | [[Image: | + | [[Image:DGAT_Mech.png|300 px|right|thumb|Figure 1: DGAT Mechanism]] |
| - | + | ||
==== DAG Binding ==== | ==== DAG Binding ==== | ||
Revision as of 18:33, 5 April 2021
Diacylglycerol acyltransferase, DGAT
| |||||||||||
References
- ↑ 1.0 1.1 Wang L, Qian H, Nian Y, Han Y, Ren Z, Zhang H, Hu L, Prasad BVV, Laganowsky A, Yan N, Zhou M. Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature. 2020 May;581(7808):329-332. doi: 10.1038/s41586-020-2280-2. Epub 2020 May, 13. PMID:32433610 doi:http://dx.doi.org/10.1038/s41586-020-2280-2
- ↑ 2.0 2.1 Sui X, Wang K, Gluchowski NL, Elliott SD, Liao M, Walther TC, Farese RV Jr. Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature. 2020 May;581(7808):323-328. doi: 10.1038/s41586-020-2289-6. Epub 2020 May, 13. PMID:32433611 doi:http://dx.doi.org/10.1038/s41586-020-2289-6
Student Contributors
- Betsy Johns
- Elise Wang
- Tyler Bihasa