User:Megan Fleshman/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 31: | Line 31: | ||
== Structural highlights == | == Structural highlights == | ||
| - | + | ACAT is a dimer of dimers, which is also known as a [https://en.wikipedia.org/wiki/Tetramer tetramer]. | |
| - | <scene name='87/877604/ | + | This <scene name='87/877604/Tetramer/2'>tetramer</scene> is about 260 kDa and is composed completely of helices, with each monomer containing 9 transmembrane helices, which have been color-coordinated to help with orientation within structures. The <scene name='87/877604/Colored_dimer/3'>dimer of ACAT</scene> was found to be the active arrangement. |
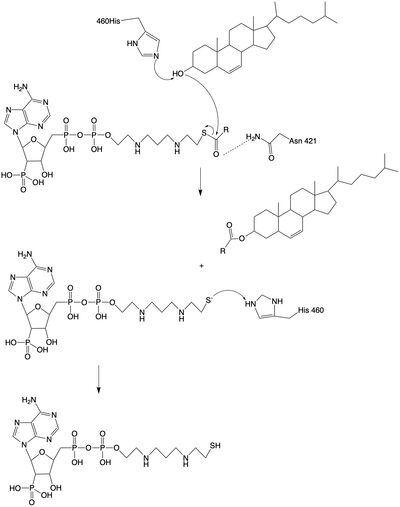
| - | <scene name='87/ | + | The <scene name='87/877604/Dimer_interface/1'>dimer-dimer interface</scene> is mobile and mostly hydrophobic, and the residues interact in a shape-complementary manner. It was also found that the reaction chamber is shielded by a lid from the cytosolic side, which leads to low catalytic activity. The binding of acyl-CoA and cholesterol induce conformational changes that activate the tunnels. Work is still being done to fully determine the mechanism of this reaction, but this is the proposed pathway. The cholesterol enters through the T tunnel while the acyl-CoA enters through the C tunnel. The reaction is catalyzed at the intersection of the two tunnels, where the His460 residue is located. The CoASH is released to the cytosol from the C tunnel, but the cholesterol ester either exits from the T tunnel to the membrane or through the L tunnel to the lumen. |
| - | The <scene name='87/ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
Revision as of 23:40, 5 April 2021
| |||||||||||
References
- ↑ Farese RV Jr. The nine lives of ACAT inhibitors. Arterioscler Thromb Vasc Biol. 2006 Aug;26(8):1684-6. doi:, 10.1161/01.ATV.0000227511.35456.90. PMID:16857957 doi:http://dx.doi.org/10.1161/01.ATV.0000227511.35456.90
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
- ↑ Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ Guan C, Niu Y, Chen SC, Kang Y, Wu JX, Nishi K, Chang CCY, Chang TY, Luo T, Chen L. Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nat Commun. 2020 May 18;11(1):2478. doi: 10.1038/s41467-020-16288-4. PMID:32424158 doi:http://dx.doi.org/10.1038/s41467-020-16288-4
Student Contributors
- Megan Fleshman, Tori Templin, Haylie Moehlenkamp