We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Megan Leaman/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 23: | Line 23: | ||
=== Mechanism === | === Mechanism === | ||
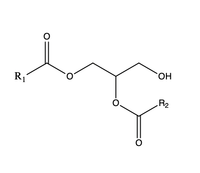
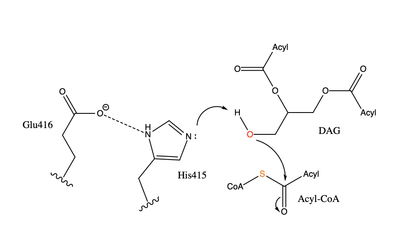
| - | [[Image:dgat chemdraw mechanism.png|400 px|right|thumb|Figure 4]]The catalytic <scene name='87/878228/His415_in_active_site/3'>His415</scene> deprotonates the hydroxyl group on the C3 of the glycerol backbone. The deprotonated oxygen then makes a nucleophilic attack on the carbonyl carbon of the Acyl-CoA, the electron density gets shifted up to the oxygen and back down to the carbonyl carbon. The sulfur of the Acyl-CoA takes the added electron density and the bond between the sulfur and carbonyl carbon is broken. Glu416 likely provides <scene name='87/878228/His415_and_glu416/2'> | + | [[Image:dgat chemdraw mechanism.png|400 px|right|thumb|Figure 4]]The catalytic <scene name='87/878228/His415_in_active_site/3'>His415</scene> deprotonates the hydroxyl group on the C3 of the glycerol backbone. The deprotonated oxygen then makes a nucleophilic attack on the carbonyl carbon of the Acyl-CoA, the electron density gets shifted up to the oxygen and back down to the carbonyl carbon. The sulfur of the Acyl-CoA takes the added electron density and the bond between the sulfur and carbonyl carbon is broken. Glu416 likely provides <scene name='87/878228/His415_and_glu416/2'>transition state stabilization</scene> of the His415 despite being outside of the 3 angstrom hydrogen binding distance. This is likely due to the fact that the DGAT 1 enzyme was unable to be visualized with the diacylglycerol in the active site. The entrance and binding of the diacylglycerol may cause conformational changes and shifting of the Glu416 to become closer to the His415. Point mutations made to His415 support the hypothesis that it is essential for stabilization in the active site since the enzyme function was completely eliminated when the mutation was made. |
=== Transition State Stabilization === | === Transition State Stabilization === | ||
Revision as of 02:24, 6 April 2021
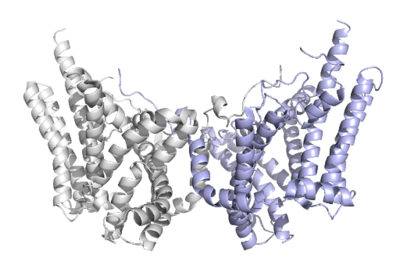
Human Diacylglycerol O-Transferase 1
| |||||||||||
References
- ↑ 1.0 1.1 Cases S, Smith SJ, Zheng YW, Myers HM, Lear SR, Sande E, Novak S, Collins C, Welch CB, Lusis AJ, Erickson SK, Farese RV Jr. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc Natl Acad Sci U S A. 1998 Oct 27;95(22):13018-23. PMID:9789033
- ↑ 2.0 2.1 Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV Jr. Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008 Nov;49(11):2283-301. doi: 10.1194/jlr.R800018-JLR200. Epub 2008, Aug 29. PMID:18757836 doi:http://dx.doi.org/10.1194/jlr.R800018-JLR200
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
- ↑ Sui, X., Wang, K., Gluchowski, N. L., Elliott, S. D., Liao, M., Walther, T. C., & Farese, R. V. (2020). Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature, 581(7808), 323-328. doi:10.1038/s41586-020-2289-6
- ↑ Wang L;Qian H;Nian Y;Han Y;Ren Z;Zhang H;Hu L;Prasad BVV;Laganowsky A;Yan N;Zhou M;. (2020, May 13). Structure and mechanism of human diacylglycerol o-acyltransferase 1. Retrieved March 09, 2021, from https://pubmed.ncbi.nlm.nih.gov/32433610/
Student Contributors
- Megan Leaman
- Grace Hall
- Karina Latsko