User:Betsy Johns/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
==Introduction== | ==Introduction== | ||
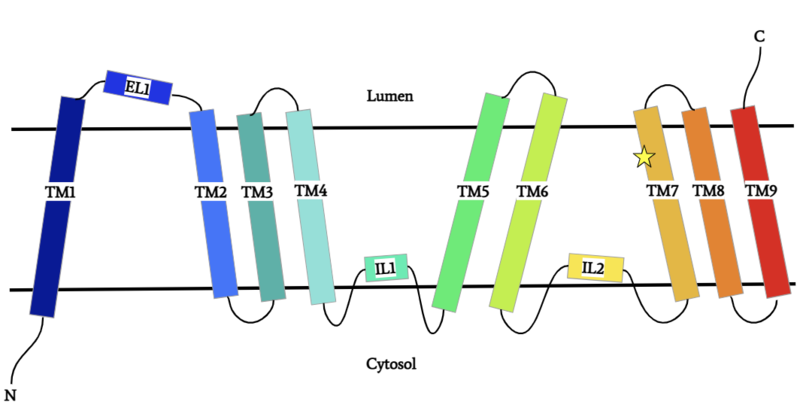
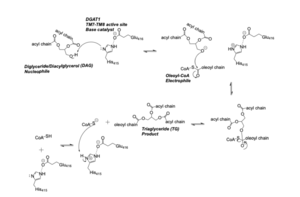
| - | Diacylglycerol acyltransferase (<scene name='87/877512/Dgat_overview/1'>DGAT | + | Diacylglycerol acyltransferase (<scene name='87/877512/Dgat_overview/1'>DGAT</scene>) is a membrane protein that synthesizes triacylglycerides from its two substrates diacylglycerol (DAG) and fatty acyl-CoA for dietary fat absorption and fat storage. DGAT synthesizes triacylglycerides from diacylglycerol (DAG) and fatty acyl-CoA. DGAT can be found expressed in the small intestine’s epithelial cells, in the liver where it synthesizes fat for storage, and in the female mammary glands where it produces fat for milk <ref name="Wang">PMID: 32433610</ref>. DGAT is a member of the membrane-bound O- acyltransferases (MBOAT) family <ref name="Sui">PMID: 32433611</ref>. All of the enzymes within this family are transmembrane enzymes that acylate lipids or proteins. MBOAT enzymes have a conserved MBOAT core, a channel-like region that acts as the enzyme’s active site. |
Revision as of 15:10, 6 April 2021
Diacylglycerol acyltransferase, DGAT
| |||||||||||
References
- ↑ 1.0 1.1 Wang L, Qian H, Nian Y, Han Y, Ren Z, Zhang H, Hu L, Prasad BVV, Laganowsky A, Yan N, Zhou M. Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature. 2020 May;581(7808):329-332. doi: 10.1038/s41586-020-2280-2. Epub 2020 May, 13. PMID:32433610 doi:http://dx.doi.org/10.1038/s41586-020-2280-2
- ↑ 2.0 2.1 Sui X, Wang K, Gluchowski NL, Elliott SD, Liao M, Walther TC, Farese RV Jr. Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature. 2020 May;581(7808):323-328. doi: 10.1038/s41586-020-2289-6. Epub 2020 May, 13. PMID:32433611 doi:http://dx.doi.org/10.1038/s41586-020-2289-6
- ↑ 3.0 3.1 Denison H, Nilsson C, Lofgren L, Himmelmann A, Martensson G, Knutsson M, Al-Shurbaji A, Tornqvist H, Eriksson JW. Diacylglycerol acyltransferase 1 inhibition with AZD7687 alters lipid handling and hormone secretion in the gut with intolerable side effects: a randomized clinical trial. Diabetes Obes Metab. 2014 Apr;16(4):334-43. doi: 10.1111/dom.12221. Epub 2013 Oct, 31. PMID:24118885 doi:http://dx.doi.org/10.1111/dom.12221
- ↑ 4.0 4.1 Stephen J, Vilboux T, Haberman Y, Pri-Chen H, Pode-Shakked B, Mazaheri S, Marek-Yagel D, Barel O, Di Segni A, Eyal E, Hout-Siloni G, Lahad A, Shalem T, Rechavi G, Malicdan MC, Weiss B, Gahl WA, Anikster Y. Congenital protein losing enteropathy: an inborn error of lipid metabolism due to DGAT1 mutations. Eur J Hum Genet. 2016 Aug;24(9):1268-73. doi: 10.1038/ejhg.2016.5. Epub 2016 Feb , 17. PMID:26883093 doi:http://dx.doi.org/10.1038/ejhg.2016.5
- ↑ Rebello CJ, Greenway FL. Obesity medications in development. Expert Opin Investig Drugs. 2020 Jan;29(1):63-71. doi:, 10.1080/13543784.2020.1705277. Epub 2019 Dec 19. PMID:31847611 doi:http://dx.doi.org/10.1080/13543784.2020.1705277
- ↑ Scott SA, Mathews TP, Ivanova PT, Lindsley CW, Brown HA. Chemical modulation of glycerolipid signaling and metabolic pathways. Biochim Biophys Acta. 2014 Aug;1841(8):1060-84. doi:, 10.1016/j.bbalip.2014.01.009. Epub 2014 Jan 15. PMID:24440821 doi:http://dx.doi.org/10.1016/j.bbalip.2014.01.009
Student Contributors
- Betsy Johns
- Elise Wang
- Tyler Bihasa