User:Kaitlyn Roberts/Sandbox 2
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
=== Tertiary Structure === | === Tertiary Structure === | ||
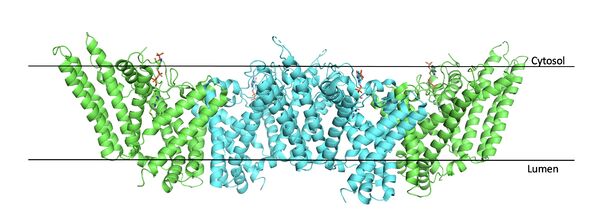
[[Image:Tetramerlabels.jpeg|600 px|right|thumb|Figure #. Tetramer within the Membrane]] | [[Image:Tetramerlabels.jpeg|600 px|right|thumb|Figure #. Tetramer within the Membrane]] | ||
| - | [[Image:Tetramer3.png|600 px|right|thumb|Figure #. Tetramer within the Membrane]] | ||
The overall structure of the enzyme is a <scene name='87/877559/Tetramer/10'>tetramer</scene> structure or a dimer of dimers. The functional building block of SOAT is a <scene name='87/877559/Dimer/3'>dimer</scene> which is made up of two identical <scene name='87/877559/Monomer/5'>monomer</scene> structures. The [https://en.wikipedia.org/wiki/Amino_acid residues] that form the dimer interface are mostly hydrophobic and interact with each other in a shape-complementary manner. Mutating residues within the dimer interface reduced the dimers to monomer fractions, indicating that the dimeric architecture is important for the activity of the enzyme. The dimerization of SOAT is mainly mediated by [https://en.wikipedia.org/wiki/Van_der_Waals_force extensive van der Waals interactions] between TM1 in one protomer and the [https://en.wikipedia.org/wiki/Lumen_(anatomy) lumenal segment] of TM6 and the [https://en.wikipedia.org/wiki/Cytosol cytosolic segment] of TM9 in the other. TM1, TM5, TM6 and TM9 from the two protomers enclose a deep hydrophobic pocket that is open to the lumenal side. Numerous hydrophobic residues on TM6 and TM9 from one protomer contact those on TM1 from the other protomer. On the intracellular side, hydrophobic residues on IH1 of each protomer interact with each other to stabilize the dimer.<ref name="Qian">PMID:32433614</ref> | The overall structure of the enzyme is a <scene name='87/877559/Tetramer/10'>tetramer</scene> structure or a dimer of dimers. The functional building block of SOAT is a <scene name='87/877559/Dimer/3'>dimer</scene> which is made up of two identical <scene name='87/877559/Monomer/5'>monomer</scene> structures. The [https://en.wikipedia.org/wiki/Amino_acid residues] that form the dimer interface are mostly hydrophobic and interact with each other in a shape-complementary manner. Mutating residues within the dimer interface reduced the dimers to monomer fractions, indicating that the dimeric architecture is important for the activity of the enzyme. The dimerization of SOAT is mainly mediated by [https://en.wikipedia.org/wiki/Van_der_Waals_force extensive van der Waals interactions] between TM1 in one protomer and the [https://en.wikipedia.org/wiki/Lumen_(anatomy) lumenal segment] of TM6 and the [https://en.wikipedia.org/wiki/Cytosol cytosolic segment] of TM9 in the other. TM1, TM5, TM6 and TM9 from the two protomers enclose a deep hydrophobic pocket that is open to the lumenal side. Numerous hydrophobic residues on TM6 and TM9 from one protomer contact those on TM1 from the other protomer. On the intracellular side, hydrophobic residues on IH1 of each protomer interact with each other to stabilize the dimer.<ref name="Qian">PMID:32433614</ref> | ||
Revision as of 14:05, 13 April 2021
Human Sterol O-acyltransferase
| |||||||||||
References
- ↑ Guan C, Niu Y, Chen SC, Kang Y, Wu JX, Nishi K, Chang CCY, Chang TY, Luo T, Chen L. Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nat Commun. 2020 May 18;11(1):2478. doi: 10.1038/s41467-020-16288-4. PMID:32424158 doi:http://dx.doi.org/10.1038/s41467-020-16288-4
- ↑ Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
Student Contributors
- Kylie Pfeifer
- Stephanie Pellegrino
- Kaitlyn Roberts