We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Madison Unger/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
== Structure == | == Structure == | ||
=== Overall Structure === | === Overall Structure === | ||
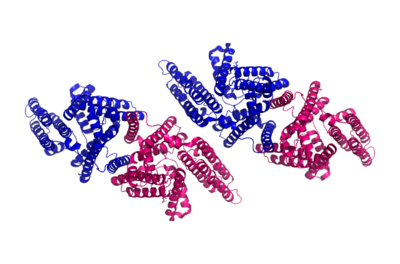
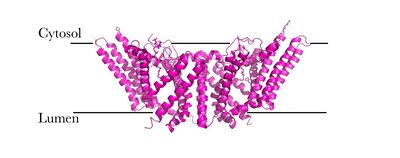
| - | ACAT is a tetramer composed of a [http://en.wikipedia.org/wiki/Protein_dimer dimer] of a dimer, but is able to perform its function solely as a dimer (Fig. 2). [[Image:Tetramer_dimer_of_dimer.png|400px|left|thumb|Figure 2: Tetrameric dimer of dimer for ACAT]]There are <scene name='87/877507/9_tm_helices_per_monomer/1'>nine transmembrane helices</scene> in each domain which create a tunnel for the active site. There are also three helices found on the intracellular side (IH1, IH2, and IH3) and one helix on the extracellular side (EH1). The active site contains three tunnels – the transmembrane tunnel for cholesterol entrance, the cytosolic tunnel for acyl-CoA entrance, and the lumen tunnel for cholesterol ester exit. ACAT also has an amino-terminal cytosolic domain (NTD) that is important for tetramerization of this protein. | + | ACAT is a tetramer composed of a [http://en.wikipedia.org/wiki/Protein_dimer dimer] of a dimer, but is able to perform its function solely as a dimer (Fig. 2). [[Image:Tetramer_dimer_of_dimer.png|400px|left|thumb|Figure 2: Tetrameric dimer of dimer for ACAT]]There are <scene name='87/877507/9_tm_helices_per_monomer/1'>nine transmembrane helices</scene> in each domain which create a tunnel for the active site. There are also three helices found on the intracellular side (IH1, IH2, and IH3) and one helix on the extracellular side (EH1). The active site contains three tunnels – the transmembrane tunnel for cholesterol entrance, the cytosolic tunnel for acyl-CoA entrance, and the lumen tunnel for cholesterol ester exit. ACAT also has an amino-terminal cytosolic domain (NTD) that is important for tetramerization of this protein.<ref name= "Cases" /> |
=== Active Site/Important Residues === | === Active Site/Important Residues === | ||
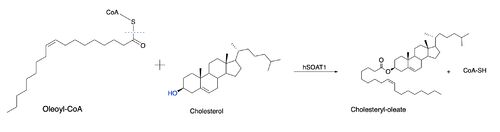
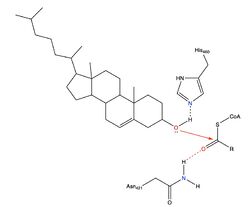
An important residue in the ACAT active site is His460, a Histidine, which is located where the tunnels converge. It is thought that His460 is located on TM7 (Qian et al.). When converting to a cholesteryl ester, the <scene name='87/877507/H460_final_1/1'>His460</scene> acts as a catalytic base that deprotonates the cholesterol. An asparagine <scene name='87/877507/Asn241/2'>Asn421</scene> is another important residue in the reaction that is able to form a hydrogen bond with acyl-CoA for stabilization. Additionally, a <scene name='87/877507/Acyl-coa_surface/2'>naturally-occurring substrate</scene> can be found in the active site of ACAT and replaced by cholesterol for synthesis of cholesteryl ester. | An important residue in the ACAT active site is His460, a Histidine, which is located where the tunnels converge. It is thought that His460 is located on TM7 (Qian et al.). When converting to a cholesteryl ester, the <scene name='87/877507/H460_final_1/1'>His460</scene> acts as a catalytic base that deprotonates the cholesterol. An asparagine <scene name='87/877507/Asn241/2'>Asn421</scene> is another important residue in the reaction that is able to form a hydrogen bond with acyl-CoA for stabilization. Additionally, a <scene name='87/877507/Acyl-coa_surface/2'>naturally-occurring substrate</scene> can be found in the active site of ACAT and replaced by cholesterol for synthesis of cholesteryl ester. | ||
Revision as of 14:49, 16 April 2021
Human Acyl-CoenzymeA
| |||||||||||
References
- ↑ Wang L, Qian H, Nian Y, Han Y, Ren Z, Zhang H, Hu L, Prasad BVV, Laganowsky A, Yan N, Zhou M. Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature. 2020 May;581(7808):329-332. doi: 10.1038/s41586-020-2280-2. Epub 2020 May, 13. PMID:32433610 doi:http://dx.doi.org/10.1038/s41586-020-2280-2
- ↑ Moorthy PS, Neelagandan K, Balasubramanian M, Ponnuswamy MN. Purification, Crystallization and Preliminary X-Ray Diffraction Studies on Goat (Capra hircus) Hemoglobin - A Low Oxygen Affinity Species. Protein Pept Lett. 2009;16(4):454-6. PMID:19356147
- ↑ 3.0 3.1 3.2 3.3 Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ 4.0 4.1 Cases S, Novak S, Zheng YW, Myers HM, Lear SR, Sande E, Welch CB, Lusis AJ, Spencer TA, Krause BR, Erickson SK, Farese RV Jr. ACAT-2, a second mammalian acyl-CoA:cholesterol acyltransferase. Its cloning, expression, and characterization. J Biol Chem. 1998 Oct 9;273(41):26755-64. doi: 10.1074/jbc.273.41.26755. PMID:9756919 doi:http://dx.doi.org/10.1074/jbc.273.41.26755
- ↑ 5.0 5.1 Guan C, Niu Y, Chen SC, Kang Y, Wu JX, Nishi K, Chang CCY, Chang TY, Luo T, Chen L. Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nat Commun. 2020 May 18;11(1):2478. doi: 10.1038/s41467-020-16288-4. PMID:32424158 doi:http://dx.doi.org/10.1038/s41467-020-16288-4
- ↑ Rogers MA, Liu J, Song BL, Li BL, Chang CC, Chang TY. Acyl-CoA:cholesterol acyltransferases (ACATs/SOATs): Enzymes with multiple sterols as substrates and as activators. J Steroid Biochem Mol Biol. 2015 Jul;151:102-7. doi: 10.1016/j.jsbmb.2014.09.008., Epub 2014 Sep 12. PMID:25218443 doi:http://dx.doi.org/10.1016/j.jsbmb.2014.09.008
- ↑ Hartmann T, Kuchenbecker J, Grimm MO. Alzheimer's disease: the lipid connection. J Neurochem. 2007 Nov;103 Suppl 1:159-70. doi: 10.1111/j.1471-4159.2007.04715.x. PMID:17986151 doi:http://dx.doi.org/10.1111/j.1471-4159.2007.04715.x
- ↑ Li J, Gu D, Lee SS, Song B, Bandyopadhyay S, Chen S, Konieczny SF, Ratliff TL, Liu X, Xie J, Cheng JX. Abrogating cholesterol esterification suppresses growth and metastasis of pancreatic cancer. Oncogene. 2016 Dec 15;35(50):6378-6388. doi: 10.1038/onc.2016.168. Epub 2016 May , 2. PMID:27132508 doi:http://dx.doi.org/10.1038/onc.2016.168
- ↑ Rudel LL, Shelness GS. Cholesterol esters and atherosclerosis-a game of ACAT and mouse. Nat Med. 2000 Dec;6(12):1313-4. doi: 10.1038/82110. PMID:11100106 doi:http://dx.doi.org/10.1038/82110
Student Contributors
- Leah Goehring
- Gabby Smith
- Anna Campbell