We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1678
From Proteopedia
(Difference between revisions)
| Line 30: | Line 30: | ||
== Other important features == | == Other important features == | ||
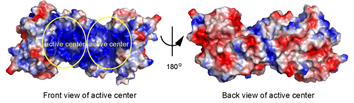
| - | As seen in the image below, the protein has two active centers since it is a dimer. In the "front view of the active center," it can be seen that each active center is made of positively charged amino acids (blue) important for binding the negatively charged substrate. | + | As seen in the image below, the protein has two active centers since it is a dimer. In the "front view of the active center," it can be seen that each active center is made of positively charged amino acids (blue) important for binding the negatively charged substrate. As seen in the "back view of the active center," the rest of the protein not containing the active site is evenly distributed with positive and negative charges because the substrate does not bind there. |
[[Image:AlyC3_active_center_positive_and_negative.png]] | [[Image:AlyC3_active_center_positive_and_negative.png]] | ||
Revision as of 18:35, 18 April 2021
| This Sandbox is Reserved from 01/25/2021 through 04/30/2021 for use in Biochemistry taught by Bonnie Hall at Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1665 through Sandbox Reserved 1682. |
To get started:
More help: Help:Editing |
Alginate Lyase (AlyC3)
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644