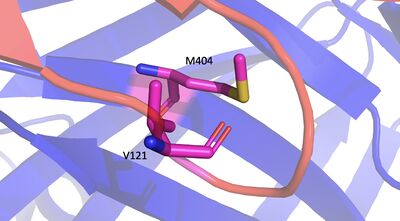
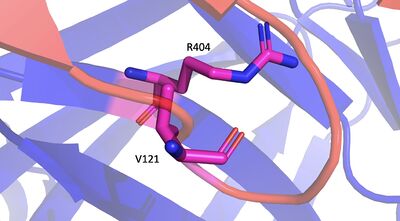
Introduction
A lipase is an enzyme that is capable of catalyzing the hydrolysis of fats/lipids which are consumed through oils. It is encoded by the p22 region in chromosome 8. Once synthesized, it is secreted into the interstitial space in several tissues. The main site of action for LPL is in the capillary lumen within muscle and adipose tissue. The function of this lipase is to hydrolyze triglycerides of very low density lipoproteins (VLDL) and to aid in the delivery of lipid nutrients to vital tissues. The enzyme is commonly found on the surface of cells that line blood capillaries. Two different lipoproteins are essential to break down triglycerides. One of the lipoproteins is utilized to transport fat into the bloodstream from different organs. The lipoproteins essential, in the transport of fat from the intestine, are referred to as chylomicrons. VLDL are utilized in carrying triglycerides from the liver into the bloodstream. The hydrolysis of triglycerides by lipoprotein lipase results in fat molecules for future usage by the body as energy or stored in fatty tissue.
Structural Overview
Lipoprotein Lipase is assumed to only be active as a , however, previous studies have argued that the lipase can be active in its monomeric form. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6442593/) The N-terminal domain of lipoprotein lipase is known to consist of an alpha/beta hydrolase domain, which is composed of six alpha helices and ten beta-strands. This domain creates an . The C-terminal domain of lipoprotein lipase is composed of twelve beta strands which form a “.
Mechanism
Lipoprotein Lipase functions to catalyze the hydrolysis of one ester bond of triglycerides. It does this by utilizing a simple serine hydrolase mechanism, in which it uses a composed of Asp183, His268, and Ser159 to catalyze the hydrolysis. His268 serves as a base catalyst by deprotonation of Ser159, which can then serve as the nucleophile. The transition state of the catalysis is stabilized by the composed of Trp82 and Leu160. The hydrolysis results in the formation of one free fatty acid and a glycerol with two fatty acid tails.
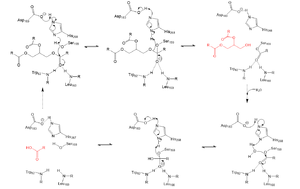
Serine hydrolase mechanism utilized by LPL to catalyze the breakdown of one ester bond of a triglyceride. Compounds colored red are the products of the hydrolysis.
Relevance & Disease
LPL is an extremely important enzyme, in that it breaks down triglycerides carried on VLDL, which leads to the reduction of cholesterol buildup. Cholesterol build up is a very serious issue with regards to obesity in the United States. In addition to this, increased plasma triglyceride levels (hypertriglyceridemia) is very unhealthy and is the leading cause of Coronary Artery Disease in America. LPL is an enzyme that helps combat this disease by breaking down the excess triglycerides that block the arteries of your heart. Very similarly, Chylomicronemia, a high level of triglycerides in the blood, causes buildup of chylomicrons (ultra-low-density lipoproteins) and leads to similar diseases. Without LPL in the body, developing coronary & metabolic (liver & pancreas) based diseases are at a higher likelihood.
Structural highlights
Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein (GPIHBP1) is necessary for LPL function and stability. (LIVE REFERENCED NEEDED)
GPI/LPL interface:
Calcium ion stabilization, lid
Calcium ion stabilization:
References
[1]
[2]
[3]
[4]
[5] (LPL GENERAL REFERENCE)
[6] (LPL GENERAL REFERENCE BOOK IF NEEDED)
Student Contributors
Giselle Flores
Dustin Soe
Maggie Stopa