We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Sarah Maarouf/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
= '''''H. sapiens'' Lipoprotein Lipase in complex with GPIHBP1 and triglyceride metabolism'''= | = '''''H. sapiens'' Lipoprotein Lipase in complex with GPIHBP1 and triglyceride metabolism'''= | ||
| - | <StructureSection load='6OB0' size='350' frame='true' side='right' caption='Lipoprotein lipase (green) bound to GPIHBP1 (cyan) (PDB | + | <StructureSection load='6OB0' size='350' frame='true' side='right' caption='Lipoprotein lipase (green) bound to GPIHBP1 (cyan) (PDB 6OB0)' scene='87/877560/General/2'> |
| Line 10: | Line 10: | ||
== Function == | == Function == | ||
| - | Energy is metabolized through the hydrolysis of triglyceride-rich lipoproteins (TRLs), among other macromolecules, to release fatty acids that can be used or stored for later energy use <ref name="Olivecrona">PMID:27031275</ref> | + | Energy is metabolized through the hydrolysis of triglyceride-rich lipoproteins (TRLs), among other macromolecules, to release fatty acids that can be used or stored for later energy use. <ref name="Olivecrona">PMID:27031275</ref> LPL is the main enzyme involved with the metabolism of triglycerides within TRLs. The LPL-GPIHBP1 complex is crucial for clearing triglycerides from the bloodstream and delivery of lipid nutrients to vital areas, such as heart and skeletal muscle, and [https://www.sciencedirect.com/topics/medicine-and-dentistry/adipose-tissue adipose tissue]. <ref name="Birrane">PMID:30559189</ref> Loss of function mutations in LPL or GPIHBP1 can have detrimental effects on the body's ability to metabolize triglycerides which in turn causes [https://medlineplus.gov/ency/article/000405.htm#:~:text=Chylomicronemia%20syndrome%20is%20a%20disorder,is%20passed%20down%20through%20families. chylomicronemia]. Elevated levels of plasma triglycerides, called hypertriglyceridemia, also increases the risk of developing [https://www.mayoclinic.org/diseases-conditions/coronary-artery-disease/symptoms-causes/syc-20350613 coronary artery disease (CAD)].<ref name=”Fong”>PMID:27185325</ref> |
== Structure == | == Structure == | ||
| Line 22: | Line 22: | ||
=== GPIHBP1 === | === GPIHBP1 === | ||
| - | GPIHBP1 is a membrane bound protein that binds to the C-terminal domain of LPL. It contains specific features, the LU domain and the acidic domain, that contribute to its binding affinity. GPIHBP1’s LU domain is 75 residues in length and adopts a <scene name='87/877554/Fingers_of_gpihbp1/2'>three-fingered fold</scene> that is stabilized by <scene name='87/877554/Disulfide_bonds_gpihbp1/5'>five disulfide bonds</scene>. The LPL-GPIHBP1 binding interface involves the three-fingered fold and depends largely on hydrophobic interactions between the two subunits. It contains a highly acidic and disordered N-terminal domain with 21 of the 26 total residues being aspartates or glutamates which were not visible in the electron density map. This indicates that the acidic domain of GPIHBP1 likely exhibits conformational flexibility<ref name="Arora">PMID:31072929</ref> | + | GPIHBP1 is a membrane bound protein that binds to the C-terminal domain of LPL. It contains specific features, the LU domain and the acidic domain, that contribute to its binding affinity. GPIHBP1’s LU domain is 75 residues in length and adopts a <scene name='87/877554/Fingers_of_gpihbp1/2'>three-fingered fold</scene> that is stabilized by <scene name='87/877554/Disulfide_bonds_gpihbp1/5'>five disulfide bonds</scene>. The LPL-GPIHBP1 binding interface involves the three-fingered fold and depends largely on hydrophobic interactions between the two subunits. It contains a highly acidic and disordered N-terminal domain with 21 of the 26 total residues being aspartates or glutamates which were not visible in the electron density map. This indicates that the acidic domain of GPIHBP1 likely exhibits conformational flexibility.<ref name="Arora">PMID:31072929</ref> |
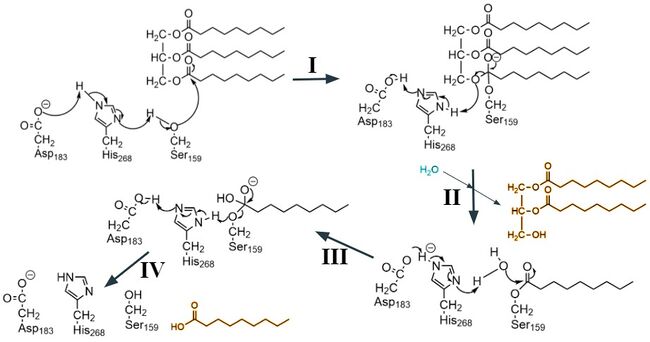
== Active Site == | == Active Site == | ||
| - | <scene name='87/877554/Active_site_residues/10'>The active site</scene> by which this enzyme catalyzes the hydrolysis of triglycerides, is located in the N-terminal domain of LPL. These residues are Ser159, Asp183, and His268<ref name="Birrane">PMID:30559189</ref> | + | <scene name='87/877554/Active_site_residues/10'>The active site</scene> by which this enzyme catalyzes the hydrolysis of triglycerides, is located in the N-terminal domain of LPL. These residues are Ser159, Asp183, and His268.<ref name="Birrane">PMID:30559189</ref> Located nearby to these residues, the <scene name='87/877554/Oxyanion_hole/9'>oxyanion hole</scene> stabilizes the transition state of the substrate through the backbone amides of Trp82 and Leu160.<ref name="Arora">PMID:31072929</ref> Surrounding the active site, is the <scene name='87/877554/Hydrophobic_binding_region/3'> hydrophobic binding region </scene> that stabilizes the hydrophobic lipid substrates in the binding pocket and forms van der Waals interactions with the hydrophobic tails of lipid that are entering. The hydrophobic region allows the substrates to access the <scene name='87/877554/Access_to_ct/3'>catalytic triad</scene> for hydrolysis.<ref name="Birrane">PMID:30559189</ref> |
=== Mechanism === | === Mechanism === | ||
| Line 70: | Line 70: | ||
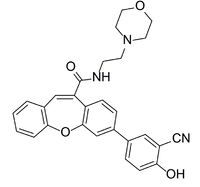
[[Image: Compound_2.jpg|200px|left|thumb|Figure 3. The structure of the inhibitor referred to as Compound 2.]] | [[Image: Compound_2.jpg|200px|left|thumb|Figure 3. The structure of the inhibitor referred to as Compound 2.]] | ||
| - | The active site residues of LPL can be blocked from preforming the breakdown of triglycerides by the presence of a a competitive inhibitor. The inhibitor binds in the hydrophobic binding region and occludes access to the catalytic triad. The inhibitor known as Compound 2 (Figure-3) was designed to stabilize the structure of the LPL/GPIHBP1 complex for the purpose of structure determination <ref name="Arora">PMID:31072929</ref> | + | The active site residues of LPL can be blocked from preforming the breakdown of triglycerides by the presence of a a competitive inhibitor. The inhibitor binds in the hydrophobic binding region and occludes access to the catalytic triad. The inhibitor known as Compound 2 (Figure-3) was designed to stabilize the structure of the LPL/GPIHBP1 complex for the purpose of structure determination.<ref name="Arora">PMID:31072929</ref> Two molecules of the <scene name='87/877554/Compound_2/1'>inhibitor</scene> fit into the hydrophobic binding region, with the proximal molecule reaching the catalytic residues where it forms interactions. |
| Line 88: | Line 88: | ||
=== Mutations === | === Mutations === | ||
| - | Missense mutations in LPL or GPIHBP1 cause a number of genetic diseases, the most prominent being [https://medlineplus.gov/ency/article/000405.htm#:~:text=Chylomicronemia%20syndrome%20is%20a%20disorder,is%20passed%20down%20through%20families. Chylomicronemia], or the inability to break down lipids. Two specific point mutations that contribute to this disease through the abolishment of LPL-GPIHBP1 binding are <scene name='87/877554/Met404_cys445/1'>M404R and C445Y</scene>. Because the LPL-GPIHBP1 complex is bound together primarily by hydrophobic interactions, switching a nonpolar [https://en.wikipedia.org/wiki/Methionine methionine] residue to a positively charged [https://en.wikipedia.org/wiki/Arginine arginine] residue, or a nonpolar [https://en.wikipedia.org/wiki/Cysteine cysteine] residue to a polar [https://en.wikipedia.org/wiki/Tyrosine tyrosine] residue will disrupt these hydrophobic interactions and weaken the interface. Also, the much larger side chain of arginine is difficult to accommodate into the hydrophobic pocket of GPIHBP1 <ref name="Birrane">PMID:30559189</ref> | + | Missense mutations in LPL or GPIHBP1 cause a number of genetic diseases, the most prominent being [https://medlineplus.gov/ency/article/000405.htm#:~:text=Chylomicronemia%20syndrome%20is%20a%20disorder,is%20passed%20down%20through%20families. Chylomicronemia], or the inability to break down lipids. Two specific point mutations that contribute to this disease through the abolishment of LPL-GPIHBP1 binding are <scene name='87/877554/Met404_cys445/1'>M404R and C445Y</scene>. Because the LPL-GPIHBP1 complex is bound together primarily by hydrophobic interactions, switching a nonpolar [https://en.wikipedia.org/wiki/Methionine methionine] residue to a positively charged [https://en.wikipedia.org/wiki/Arginine arginine] residue, or a nonpolar [https://en.wikipedia.org/wiki/Cysteine cysteine] residue to a polar [https://en.wikipedia.org/wiki/Tyrosine tyrosine] residue will disrupt these hydrophobic interactions and weaken the interface. Also, the much larger side chain of arginine is difficult to accommodate into the hydrophobic pocket of GPIHBP1. <ref name="Birrane">PMID:30559189</ref> These mutations do not affect LPL’s activity or secretion but affect its ability to be transported to its site of action in the capillary lumen. This leads to an accumulation of unmetabolized lipids but also catalytically active LPL in the interstitial spaces.<ref name=”Voss”>PMID:21518912</ref> |
| - | Additionally, <scene name='87/877554/Mutation_of_d202_and_d201/3'>the point mutations p.D201V and p.D202E</scene> observed in patients with [https://medlineplus.gov/ency/article/000405.htm#:~:text=Chylomicronemia%20syndrome%20is%20a%20disorder,is%20passed%20down%20through%20families. Chylomicronemia] has been observed to eliminate LPL secretion and reduce its activity. Among other residues, the carboxylic acid side chain of D201 is critical for coordinating LPL’s calcium ion. Mutating this negatively charged [https://en.wikipedia.org/wiki/Aspartic_acid aspartic acid] into a nonpolar [https://en.wikipedia.org/wiki/Valine valine] residue disrupts the ionic bond between calcium and the aspartate, disturbing the overall calcium binding. A similar mutation has also been observed for p.D202E. These two mutations, in turn, destabilize LPL folding and thereby prevent its secretion from cells<ref name="Birrane">PMID:30559189</ref> | + | Additionally, <scene name='87/877554/Mutation_of_d202_and_d201/3'>the point mutations p.D201V and p.D202E</scene> observed in patients with [https://medlineplus.gov/ency/article/000405.htm#:~:text=Chylomicronemia%20syndrome%20is%20a%20disorder,is%20passed%20down%20through%20families. Chylomicronemia] has been observed to eliminate LPL secretion and reduce its activity. Among other residues, the carboxylic acid side chain of D201 is critical for coordinating LPL’s calcium ion. Mutating this negatively charged [https://en.wikipedia.org/wiki/Aspartic_acid aspartic acid] into a nonpolar [https://en.wikipedia.org/wiki/Valine valine] residue disrupts the ionic bond between calcium and the aspartate, disturbing the overall calcium binding. A similar mutation has also been observed for p.D202E. These two mutations, in turn, destabilize LPL folding and thereby prevent its secretion from cells.<ref name="Birrane">PMID:30559189</ref> |
Revision as of 16:37, 20 April 2021
H. sapiens Lipoprotein Lipase in complex with GPIHBP1 and triglyceride metabolism
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Arora R, Nimonkar AV, Baird D, Wang C, Chiu CH, Horton PA, Hanrahan S, Cubbon R, Weldon S, Tschantz WR, Mueller S, Brunner R, Lehr P, Meier P, Ottl J, Voznesensky A, Pandey P, Smith TM, Stojanovic A, Flyer A, Benson TE, Romanowski MJ, Trauger JW. Structure of lipoprotein lipase in complex with GPIHBP1. Proc Natl Acad Sci U S A. 2019 May 21;116(21):10360-10365. doi:, 10.1073/pnas.1820171116. Epub 2019 May 9. PMID:31072929 doi:http://dx.doi.org/10.1073/pnas.1820171116
- ↑ Olivecrona G. Role of lipoprotein lipase in lipid metabolism. Curr Opin Lipidol. 2016 Jun;27(3):233-41. doi: 10.1097/MOL.0000000000000297. PMID:27031275 doi:http://dx.doi.org/10.1097/MOL.0000000000000297
- ↑ 3.0 3.1 3.2 3.3 3.4 Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116
- ↑ Fong LG, Young SG, Beigneux AP, Bensadoun A, Oberer M, Jiang H, Ploug M. GPIHBP1 and Plasma Triglyceride Metabolism. Trends Endocrinol Metab. 2016 Jul;27(7):455-469. doi: 10.1016/j.tem.2016.04.013. , Epub 2016 May 14. PMID:27185325 doi:http://dx.doi.org/10.1016/j.tem.2016.04.013
- ↑ Voss CV, Davies BS, Tat S, Gin P, Fong LG, Pelletier C, Mottler CD, Bensadoun A, Beigneux AP, Young SG. Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proc Natl Acad Sci U S A. 2011 May 10;108(19):7980-4. doi:, 10.1073/pnas.1100992108. Epub 2011 Apr 25. PMID:21518912 doi:http://dx.doi.org/10.1073/pnas.1100992108
Student Contributors
- Aniyah Coles
- Sarah Maarouf
- Audrey Marjamaa