User:Hannah Wright/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
==Introduction== | ==Introduction== | ||
| - | + | ||
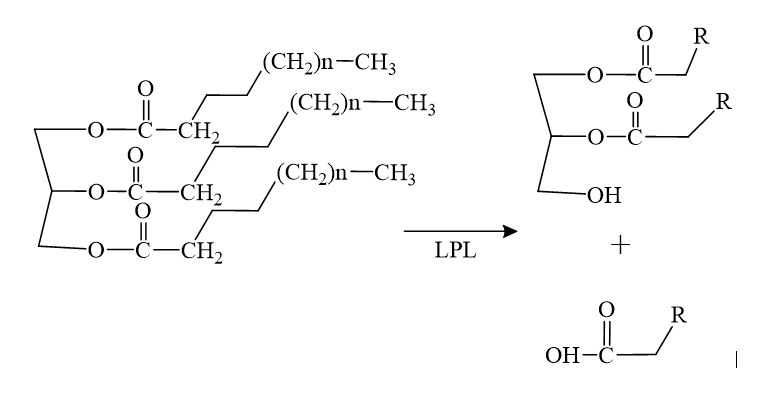
| - | LPL, lipoprotein lipase, is an enzyme that affects the breakdown of triglycerides which are carried from various organs to the blood by molecules called lipoproteins. LPL is found on the surface of cells lining the capillaries within muscles and fatty tissue. | + | LPL, '''lipoprotein lipase''', is an enzyme that affects the breakdown of triglycerides which are carried from various organs to the blood by molecules called lipoproteins. LPL is found on the surface of cells lining the capillaries within muscles and fatty tissue. |
[[Image:Triglyceride.png]] | [[Image:Triglyceride.png]] | ||
| - | LPL was identified more than 60 years ago and studied by biochemists and physiologists intensely since. it wasn’t until recently that LPL’s detailed structure was determined due to LPL’s hydrolase domain susceptibility to unfolding. LMF1 and GPIHBP1, glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1 ,were discovered to be required for proper folding and enzymatic activity of LPL. LMF1, lipase maturation factor 1, is a chaperone protein that is responsible for proper folding and secretion of LPL. Through the use of X-ray Crystallography it was also discovered that LPL is a monomer rather than the previously believed homodimer. | + | LPL was identified more than 60 years ago and studied by biochemists and physiologists intensely since. it wasn’t until recently that LPL’s detailed structure was determined due to LPL’s hydrolase domain susceptibility to unfolding. LMF1 and GPIHBP1, '''glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1''' ,were discovered to be required for proper folding and enzymatic activity of LPL. LMF1,''' lipase maturation factor 1''', is a chaperone protein that is responsible for proper folding and secretion of LPL. Through the use of X-ray Crystallography it was also discovered that LPL is a monomer rather than the previously believed homodimer. |
===Function=== | ===Function=== | ||
| Line 18: | Line 18: | ||
===Overall Structure=== | ===Overall Structure=== | ||
Overall, LPL is a monomer but biologically it is paired with another monomer. These are often referred two as <scene name='87/877603/Chain_a_and_chain_b/2'>Chain A and Chain B</scene>. Chain A (shown as slate blue) and Chain B (shown as magenta) are identical besides orientation. Chain A and B are oriented head to tail. LPL is also complexed with GPIHBP1 (shown as cyan) and is essential for LPL to remain stable and avoid denaturation | Overall, LPL is a monomer but biologically it is paired with another monomer. These are often referred two as <scene name='87/877603/Chain_a_and_chain_b/2'>Chain A and Chain B</scene>. Chain A (shown as slate blue) and Chain B (shown as magenta) are identical besides orientation. Chain A and B are oriented head to tail. LPL is also complexed with GPIHBP1 (shown as cyan) and is essential for LPL to remain stable and avoid denaturation | ||
| - | ===LPL===LPL has two main domains, the larger N-terminus domain containing the active site and the smaller C-terminus domain. These two domains are connected via a peptide linker hinge. LPL also contains a large basic patch and a single calcium ion. Additionally, LPL consists of two <scene name='87/877603/Glycans/3'>N-linked glycans</scene> which likely contributes to the correct folding of LPL due to the attached oligosaccharides. Five disulfide bonds contribute to the stabilization throughout LPL’s structure. | + | ===LPL===LPL has two main domains, the larger N-terminus domain containing the active site and the smaller C-terminus domain. These two domains are connected via a peptide linker hinge. LPL also contains a large basic patch and a single calcium ion. Additionally, LPL consists of two <scene name='87/877603/Glycans/3'>N-linked glycans</scene> which likely contributes to the correct folding of LPL due to the attached oligosaccharides. Five disulfide bonds contribute to the stabilization throughout LPL’s structure. Lastly, the active site in the larger N-terminus domain is lined with hydrophobic residues. |
===GPIHBP1=== | ===GPIHBP1=== | ||
GPIHBP1 (glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1) is a three-fingered LU (Ly6/uPAR) domain. GPIHBP1 is stabilized by five disulfide bonds and is also known for its N-terminal intrinsically disordered acidic domain. | GPIHBP1 (glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1) is a three-fingered LU (Ly6/uPAR) domain. GPIHBP1 is stabilized by five disulfide bonds and is also known for its N-terminal intrinsically disordered acidic domain. | ||
| Line 31: | Line 31: | ||
he calcium ion has been shown to convert inactive LPL to the active dimer form. The calcium ion is coordinated by residues A194, R197, S199, D201, and D202. Mutations in the side chain of D201, for example, can give rise to detrimental metabolic diseases as LPL can no longer | he calcium ion has been shown to convert inactive LPL to the active dimer form. The calcium ion is coordinated by residues A194, R197, S199, D201, and D202. Mutations in the side chain of D201, for example, can give rise to detrimental metabolic diseases as LPL can no longer | ||
===Active Site=== | ===Active Site=== | ||
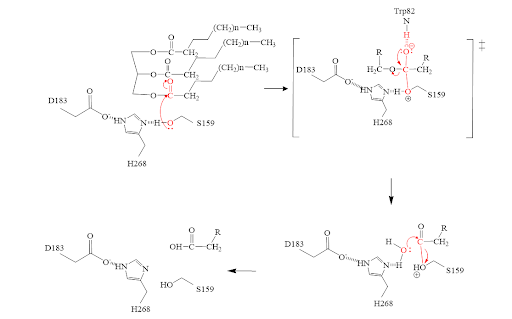
| - | The <scene name='87/878236/ | + | The <scene name='87/878236/Active_site_done/4'>active site</scene> of LPL is composed of multiple pieces. The <scene name='87/878236/Active_site_hphob_fixed/7'>hydrophobic entry</scene> to the binding site outlines the general structure and provides considerable stability to the active site. The <scene name='87/878236/Active_site_lid_region_fixed/3'>lid region</scene> is also an important component of the active site, occupying residues 243-266, which is vital for the recognition of substrates. The <scene name='87/878236/Active_site_w_catalytic_residu/4'>catalytic residues</scene> are H268, S159, and D183 which catalyze the reaction of the typical substrate of LPL, triglycerides. The oxyanion hole, consisting of residues L160, and W82, aids in the overall stability of the active site and the transition state of substrates. The main chain nitrogens stabilize the tetrahedral intermediate. |
====Mechanism==== | ====Mechanism==== | ||
Revision as of 18:51, 20 April 2021
Lipoprotein Lipase (LPL) complexed with GPIHBP1
| |||||||||||
References
- ↑ Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116
Student/Contributors
- Ashrey Burely
- Allison Welz
- Hannah Wright