Introduction
Diacylglycerol acyltransferase () is a transmembrane protein that synthesizes triacylglyceridesfrom its two substrates diacylglycerol (DAG) and fatty acyl-CoAfor dietary fat absorption and fat storage [1]. DGAT can be found expressed in the epithelial cells of the small intestine, in the liver where it synthesizes fat for storage, and in the female mammary glands where it produces fat for milk [1]. DGAT is a member of the membrane-bound O-acyltransferase (MBOAT) family [2]. All of the enzymes within this MBOAT family are transmembrane enzymes that acylate lipids or proteins [3]. MBOAT enzymes have a conserved MBOAT core, which is a channel-like region that acts as the enzyme’s active site [3]. Additionally, enzymes within the MBOAT family also contain a catalytic histidine within the active site [3].
Structure

Figure 1: DGAT Structure Overview Shown is one transmembrane subunit of DGAT with its N-terminus on the cytosolic side and its C-terminus on the luminal side. Labeled are the transmembrane domains (TM 1-7), the intracellular loops (IL1 and IL2), and the ER lumenal loop (EL1). The catalytic Histidine (His415) is labeled with a star on TM7.
DGAT is a dimer that has two identical . Each of the individual subunits contains an MBOAT core that acts as its active site. Each subunit also contains (TM), 2 intracellular loops (IL), and one ER lumenal loop (EL). TM2-9, IL1, and IL2 form the structure of the MBOAT core active site. A schematic of DGAT’s structure is shown in Figure 1.
Dimer Interface
DGAT is a dimer that has two identical subunits with 9 transmembrane alpha helices (TM), 2 intracellular loops (IL), and one ER luminal loop (EL). The dimer is held together at the . Both between the residues of the TM1 and EL1 regions of both subunits act to hold the subunits of the dimer together.
Active Site
Depiction of the DGAT active site is shown in Figure 2. Oleoyl-CoA,the 18 carbon chain version of Acyl-CoA, was used by both of the substrate-bound PDB files. DAG enters the active site of DGAT through the lateral gate, while the catalytic His415 undergoes a conformational change to widen the cytosolic channel opening so Oleoyl-CoA can enter. When the Oleoyl-CoA and DAG are within close proximity in the active site, the catalytic His415 catalyzes the reaction. The CoA head group leaves back through the channel located on the cytosolic side, while the product triacylglyceride leaves back through the lateral gate.
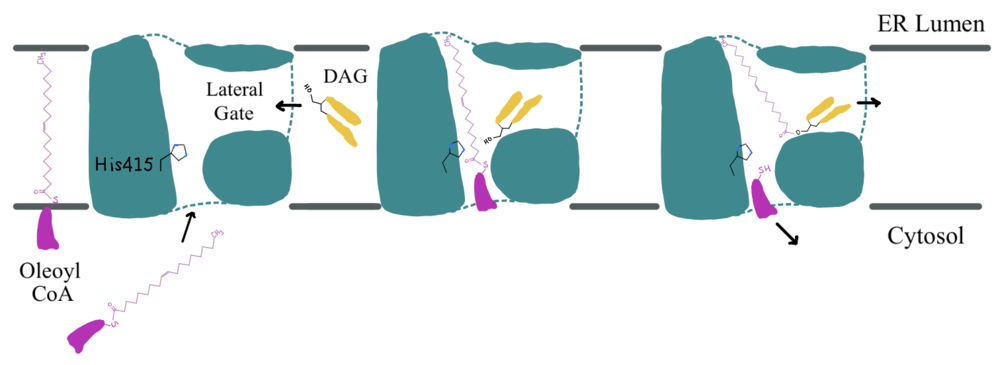
Figure 2: Active Site Mechanism Overview Modeled is the active site of DGAT (shown in teal) with its catalytic Histidine (His415), Oleoyl CoA (shown in pink), and a general diacylglycerol (DAG, shown in yellow). DAG enters the active site of DGAT through its lateral gate while the catalytic His415 flips from the cytosolic side to the lumenal side, in order for the channel opening located on the cytosolic side of DGAT to widen enough to accommodate the Oleoyl-CoA. The Oleoyl CoA then enters the active site through the channel opening located on the cytosolic side of DGAT. When the Oleoyl-CoA and DAG are within close proximity in the active site, the catalytic His415 catalyzes the reaction. The CoA head group leaves back through the channel located on the cytosolic side, while the product triacylglyceride leaves back through the lateral gate.
Oleoyl-CoA Binding
enters DGAT’s active site through a channel on the cytosolic side of the membrane. In order for the channel to accommodate the fatty acid tail of the Acyl-CoA, His415 must first break its interactions with Met434. This allows for the His415 to swing down and have hydrogen bond interactions with Gln465, thus widening the channel enough for the fatty acid tail to move into the active site. Once Acyl CoA is bound in the active site, residues Asn378, Gln437, Met434, His415, and Gln465 directly interact with and stabilize the fatty acid tail within the cytosolic channel of the active site.
DAG Binding
enters the active site through the lateral gate located in the lipid bilayer of the membrane. This lateral gate is a bent and hydrophobic channel that allows for hydrophobic linear or curvilinear molecules to enter [2]. The lateral gate channel is designed to allow for the entrance of DAG and the exit of a triacylglyceride. This channel is also lined with Phe342, Leu261, Val381, and Asn378. Once within the channel, DAG is positioned in close proximity to the bound Acyl-CoA and the catalytic His415.
Active Site Structure
The is located within the pocket formed from TM2-TM9, IL1, and IL2, with the catalytic residue Histidine 415 located on TM7. It is accessible through openings both on the cytosolic and luminal sides. The active site of DGAT serves its catalytic function by placing the His415 residue in close proximity to the acyl-CoA in order to cleave its ester bond and bind the fatty acid to the diacylglycerol. The conserved His415 acts catalytically by a charge relay system, where the negative charge of the neighboring Glu416 pulls on the electrons of histidine at the N1 position, making the N3 position more nucleophilic. This nitrogen will then deprotonate DAG so it can begin its attack on Acyl-CoA through acyl substitution. The catalytic mechanism for DGAT is shown in Figure 3 [1].
The oxyanion hole of the acyl substitution intermediate should be stabilized by DGAT. However, the two available PDB files for the substrate-bound DGAT do not show a possible stabilizing residue for the Oleoyl-CoA, the 18 carbon chain version of Acyl-CoA. The PDB file shows the sulfur of Oleoyl-CoA, the electrophile of the mechanism, facing away from His415. This is not likely to be the proper orientation for Oleoyl-CoA, as this sulfur would have to be facing towards the His415 to be protonated after the acyl substitution. For an oxyanion stabilizing residue to be located within this PDB file, the Oleyl-CoA would have to be properly oriented. The other substrate-bound PDB file, , shows the sulfur of Oleyl-CoA facing the His415. However, the tail of Oleyl-CoA is curved around the oxyanion hole, preventing a stabilizing residue within DGAT to be found. For these reasons, an oxyanion stabilizing residue was not identified in the mechanism nor in the images.
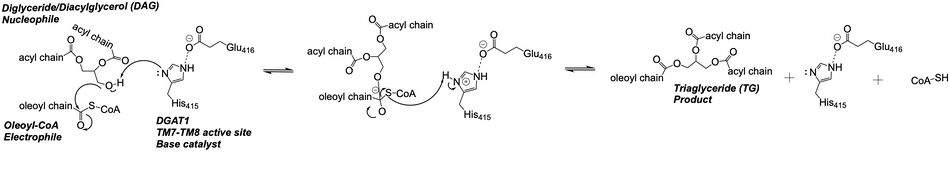
Figure 3: DGAT Mechanism The DGAT mechanism is an acyl substitution with DAG as the nucleophile and Acyl-CoA as the electrophile. His415 is the catalytic residue that deprotonates DAG to make it a better nucleophile. Glu416 stabilizes the His415. The residue that stabilizes the oxyanion hole of the intermediate is unknown.
Disease
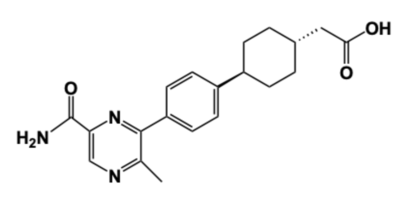
Figure 4: DGAT Inhibitor AZD7687 Shown is the structure of AZD7687, a known inhibitor of DGAT.
Obesity and nonalcoholic fatty liver disease (NAFLD) result from an accumulation of triglycerides within the body. Recently, DGAT has become a therapeutic target for obesity and nonalcoholic fatty liver disease in order to reduce triglyceride storage within the body. Different inhibitors have been created, such as AstraZeneca’s direct inhibitor AZD7687, shown in Figure 4 [4]. AZD7687 has an EC50 value 0.44 µmol/L, showing that it binds with high affinity at a low concentration of DGAT. However, while triglyceride accumulation decreased, negative side effects did occur, such as diarrhea and other adverse GI symptoms.
Additionally, congenital protein-losing enteropathy (PLE) is linked to DGAT mutations. PLE is a GI disorder that causes malabsorption of fat and a deficiency in fat-soluble vitamins. Patients in a congenital PLE case study exhibited a homozygous missense Leu295Pro mutation within the MBOAT core of their DGAT enzymes [5]. is located within the MBOAT core active site on TM5. While the Leu295 is not near the catalytic residues His415 and Glu416, the mutation will disrupt the overall active site. Proline is an alpha helix breaker because it causes steric hindrance within the backbone of the helix turn. It is hypothesized that this mutation breaks this helix in the MBOAT core and greatly reduces its enzymatic activity and ability to make triacylglycerides. Without proper DGAT function to produce triacylglycerides, there is a decrease in albumin, which is a protein that helps prevent fluid from leaking out of the liver and blood vessels. This decrease in albumin then leads to decreased efficiency in nutrient transport and fat absorption.
Relevance
[1]
[2]
[5]
[4]
[3]




