User:Sarah Maarouf/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 24: | Line 24: | ||
=== GPIHBP1 === | === GPIHBP1 === | ||
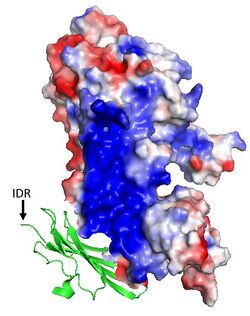
| - | [[Image: Electro.jpg| | + | [[Image: Electro.jpg|250px|right|thumb|Figure 2. The intrinsically disordered region (IDR) of GPIHBP1's N-terminal domain interacts with LPL's basic patch (blue) to stabilize LPL structure and activity.]] |
| - | GPIHBP1 is a membrane bound protein that binds to the C-terminal domain of LPL. It contains specific features, the cysteine-rich LU domain and an intrinsically disordered acidic N-terminal domain, that contribute to its binding affinity of LPL. GPIHBP1’s [https://en.wikipedia.org/wiki/LU_domain#:~:text=The%20LU%20domain%20(Ly%2D6,CD59%2C%20and%20Sgp%2D2. LU domain (Ly6-uPar)] is 75 residues in length and adopts a <scene name='87/877554/Fingers_of_gpihbp1/3'>three-fingered fold</scene>, characteristic of the LU protein family, that is stabilized by <scene name='87/877554/Disulfide_bonds_gpihbp1/6'>five disulfide bonds</scene>. The LPL-GPIHBP1 binding interface involves the three-fingered fold and depends largely on hydrophobic interactions between the two subunits, that creates a high binding affinity of LPL. The disordered acidic N-terminal domain of GPIHBP1 plays a role in being able to capture LPL also contributing to binding affinity.<ref name = "Fong">PMID: 27185325</ref> The disordered N-terminal domain has 21 of the 26 total residues being aspartates or glutamates. Unfortunately images for the acidic domain were not resolved, indicating that it likely exhibits conformational flexibility.<ref name="Arora">PMID:31072929</ref> GPIHBP1 makes electrostatic interactions with LPL's basic patch (Figure 2), | + | GPIHBP1 is a membrane bound protein that binds to the C-terminal domain of LPL. It contains specific features, the cysteine-rich LU domain and an intrinsically disordered acidic N-terminal domain, that contribute to its binding affinity of LPL. GPIHBP1’s [https://en.wikipedia.org/wiki/LU_domain#:~:text=The%20LU%20domain%20(Ly%2D6,CD59%2C%20and%20Sgp%2D2. LU domain (Ly6-uPar)] is 75 residues in length and adopts a <scene name='87/877554/Fingers_of_gpihbp1/3'>three-fingered fold</scene>, characteristic of the LU protein family, that is stabilized by <scene name='87/877554/Disulfide_bonds_gpihbp1/6'>five disulfide bonds</scene>. The LPL-GPIHBP1 binding interface involves the three-fingered fold and depends largely on hydrophobic interactions between the two subunits, that creates a high binding affinity of LPL. The disordered acidic N-terminal domain of GPIHBP1 plays a role in being able to capture LPL also contributing to binding affinity.<ref name = "Fong">PMID: 27185325</ref> The disordered N-terminal domain has 21 of the 26 total residues being aspartates or glutamates. Unfortunately images for the acidic domain were not resolved, indicating that it likely exhibits conformational flexibility.<ref name="Arora">PMID:31072929</ref> GPIHBP1 makes electrostatic interactions with LPL's basic patch (Figure 2), this complex between GPIHBP1's acidic N-terminal domain and LPL's C-terminal domain is speculated to create the stability in LPL that allows for its catalytic activity to continue when bound to GPIHBP1.<ref name="Birrane">PMID:30559189</ref> |
Revision as of 16:06, 23 April 2021
H. sapiens Lipoprotein Lipase in complex with GPIHBP1 and triglyceride metabolism
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Fong LG, Young SG, Beigneux AP, Bensadoun A, Oberer M, Jiang H, Ploug M. GPIHBP1 and Plasma Triglyceride Metabolism. Trends Endocrinol Metab. 2016 Jul;27(7):455-469. doi: 10.1016/j.tem.2016.04.013. , Epub 2016 May 14. PMID:27185325 doi:http://dx.doi.org/10.1016/j.tem.2016.04.013
- ↑ 2.0 2.1 Voss CV, Davies BS, Tat S, Gin P, Fong LG, Pelletier C, Mottler CD, Bensadoun A, Beigneux AP, Young SG. Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proc Natl Acad Sci U S A. 2011 May 10;108(19):7980-4. doi:, 10.1073/pnas.1100992108. Epub 2011 Apr 25. PMID:21518912 doi:http://dx.doi.org/10.1073/pnas.1100992108
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Arora R, Nimonkar AV, Baird D, Wang C, Chiu CH, Horton PA, Hanrahan S, Cubbon R, Weldon S, Tschantz WR, Mueller S, Brunner R, Lehr P, Meier P, Ottl J, Voznesensky A, Pandey P, Smith TM, Stojanovic A, Flyer A, Benson TE, Romanowski MJ, Trauger JW. Structure of lipoprotein lipase in complex with GPIHBP1. Proc Natl Acad Sci U S A. 2019 May 21;116(21):10360-10365. doi:, 10.1073/pnas.1820171116. Epub 2019 May 9. PMID:31072929 doi:http://dx.doi.org/10.1073/pnas.1820171116
- ↑ Young SG, Fong LG, Beigneux AP, Allan CM, He C, Jiang H, Nakajima K, Meiyappan M, Birrane G, Ploug M. GPIHBP1 and Lipoprotein Lipase, Partners in Plasma Triglyceride Metabolism. Cell Metab. 2019 Jul 2;30(1):51-65. doi: 10.1016/j.cmet.2019.05.023. PMID:31269429 doi:http://dx.doi.org/10.1016/j.cmet.2019.05.023
- ↑ Olivecrona G. Role of lipoprotein lipase in lipid metabolism. Curr Opin Lipidol. 2016 Jun;27(3):233-41. doi: 10.1097/MOL.0000000000000297. PMID:27031275 doi:http://dx.doi.org/10.1097/MOL.0000000000000297
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116
Student Contributors
- Aniyah Coles
- Sarah Maarouf
- Audrey Marjamaa