We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Kaitlyn Roberts/Sandbox 2
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
== Introduction == | == Introduction == | ||
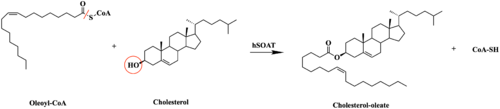
| - | [[Image:SOATfirstreaction.png|500 px|right|thumb|Figure 1. Esterification Reaction of Oleoyl-CoA and Cholesterol catalyzed by SOAT]] Sterol O-acyltransferase(SOAT), otherwise known as Acyl-coenzyme A:cholesterol acyltransferase(ACAT), is the first discovered member of the membrane-bound O-acyl [https://en.wikipedia.org/wiki/Transferase transferase] or MBOAT enzyme group. MBOAT enzymes are responsible for the transfer of [https://en.wikipedia.org/wiki/Acyl_group acyl chains] onto multiple types of substrates within the cell. There are 11 MBOAT enzyme types that can be found in humans, all of which serve a different function in the overall makeup of human biology.<ref name="Guan">PMID:32424158</ref> | + | [[Image:SOATfirstreaction.png|500 px|right|thumb|'''Figure 1.''' Esterification Reaction of Oleoyl-CoA and Cholesterol catalyzed by SOAT]] Sterol O-acyltransferase(SOAT), otherwise known as Acyl-coenzyme A:cholesterol acyltransferase(ACAT), is the first discovered member of the membrane-bound O-acyl [https://en.wikipedia.org/wiki/Transferase transferase] or MBOAT enzyme group. MBOAT enzymes are responsible for the transfer of [https://en.wikipedia.org/wiki/Acyl_group acyl chains] onto multiple types of substrates within the cell. There are 11 MBOAT enzyme types that can be found in humans, all of which serve a different function in the overall makeup of human biology.<ref name="Guan">PMID:32424158</ref> |
SOAT specifically catalyzes the [https://en.wikipedia.org/wiki/Fischer–Speier_esterification esterification] of cholesterol for efficient storage within the cell. [https://en.wikipedia.org/wiki/Cholesterol Cholesterol] is a type of membrane lipid that is responsible for controlling the fluidity and integrity of the membrane, along with other important biological processes. When there are high concentrations of cholesterol in the cell, [https://en.wikipedia.org/wiki/Cholesteryl_ester cholesteryl esters] can be formed for storage within the membrane.<ref name="Guan" /> | SOAT specifically catalyzes the [https://en.wikipedia.org/wiki/Fischer–Speier_esterification esterification] of cholesterol for efficient storage within the cell. [https://en.wikipedia.org/wiki/Cholesterol Cholesterol] is a type of membrane lipid that is responsible for controlling the fluidity and integrity of the membrane, along with other important biological processes. When there are high concentrations of cholesterol in the cell, [https://en.wikipedia.org/wiki/Cholesteryl_ester cholesteryl esters] can be formed for storage within the membrane.<ref name="Guan" /> | ||
| Line 9: | Line 9: | ||
== Structure == | == Structure == | ||
=== Tertiary Structure === | === Tertiary Structure === | ||
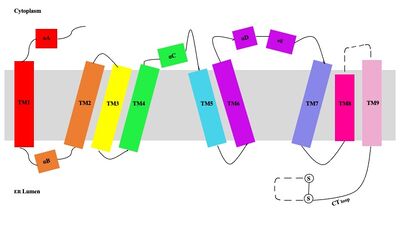
| - | [[Image:Tetramerlabels.jpeg|450 px|right|thumb|Figure 2. Tetramer unit of SOAT shown with membrane markers]] The overall structure of the enzyme is a <scene name='87/877559/Tetramer/10'>tetramer</scene> structure or a dimer of dimers. The functional building block of SOAT is a <scene name='87/877559/Dimer/3'>dimer</scene> which is made up of two identical <scene name='87/877559/Monomer/5'>monomer</scene> units. The residues that form the dimer interface are mostly hydrophobic and interact with each other in a shape-complementary manner. Mutating residues within the dimer interface reduced the dimers to monomer fractions, indicating that the dimeric architecture is important for the activity of the enzyme. Each monomer is organized into 9 <scene name='87/877559/Helices_1-9/4'>transmembrane helices</scene>. The dimerization of SOAT is mainly mediated by extensive [https://en.wikipedia.org/wiki/Van_der_Waals_force van der Waals interactions] between TM1 in one protomer and the [https://en.wikipedia.org/wiki/Lumen_(anatomy) lumenal segment] of TM6 and the [https://en.wikipedia.org/wiki/Cytosol cytosolic segment] of TM9 in the other. TM1, TM5, TM6 and TM9 from the two protomers enclose a deep hydrophobic pocket that is open to the lumenal side. Numerous hydrophobic residues on TM6 and TM9 from one protomer contact those on TM1 from the other protomer. On the intracellular side, hydrophobic residues on IH1 of each protomer interact with each other to stabilize the dimer. <ref name="Qian">PMID:32433614</ref> | + | [[Image:Tetramerlabels.jpeg|450 px|right|thumb|'''Figure 2.''' Tetramer unit of SOAT shown with membrane markers]] The overall structure of the enzyme is a <scene name='87/877559/Tetramer/10'>tetramer</scene> structure or a dimer of dimers. The functional building block of SOAT is a <scene name='87/877559/Dimer/3'>dimer</scene> which is made up of two identical <scene name='87/877559/Monomer/5'>monomer</scene> units. The residues that form the dimer interface are mostly hydrophobic and interact with each other in a shape-complementary manner. Mutating residues within the dimer interface reduced the dimers to monomer fractions, indicating that the dimeric architecture is important for the activity of the enzyme. Each monomer is organized into 9 <scene name='87/877559/Helices_1-9/4'>transmembrane helices</scene>. The dimerization of SOAT is mainly mediated by extensive [https://en.wikipedia.org/wiki/Van_der_Waals_force van der Waals interactions] between TM1 in one protomer and the [https://en.wikipedia.org/wiki/Lumen_(anatomy) lumenal segment] of TM6 and the [https://en.wikipedia.org/wiki/Cytosol cytosolic segment] of TM9 in the other. TM1, TM5, TM6 and TM9 from the two protomers enclose a deep hydrophobic pocket that is open to the lumenal side. Numerous hydrophobic residues on TM6 and TM9 from one protomer contact those on TM1 from the other protomer. On the intracellular side, hydrophobic residues on IH1 of each protomer interact with each other to stabilize the dimer. <ref name="Qian">PMID:32433614</ref> |
| - | [[Image:Helicesdiagram1.jpeg|400 px|right|thumb|Figure 3. Labeled helices of SOAT within the membrane]] | + | [[Image:Helicesdiagram1.jpeg|400 px|right|thumb|'''Figure 3.''' Labeled helices of SOAT within the membrane]] |
=== Tunnel System === | === Tunnel System === | ||
| - | [[Image:Tunnels2.jpg|250 px|right|thumb|Figure 4. Tunnel system of SOAT]] | + | [[Image:Tunnels2.jpg|250 px|right|thumb|'''Figure 4.''' Tunnel system of SOAT]] |
A main structural element of this enzyme is the tunnel systems. There are 3 main tunnels in each monomer: the cytosolic (C) tunnel opening to the cytosol, the transmembrane(T) tunnel opening to the membrane, and the lumenal(L) tunnel opens to the lumen. The C tunnel opens to the cytosol of the cell and is the entrance site for the Acyl CoA into the active site. Surface representations of SOAT indicate that there are 2 alpha helices that block the entrance to the C tunnel, therefore a conformational change needs to occur to move the 2 helices so the substrate can enter the tunnel. The T tunnel opens into the membrane and is where cholesterol enters to have access to the active site. The two substrates are catalyzed by the H460 in the active site to form the cholesteryl ester. The products then leave via different pathways. The CoA-SH in the C tunnel leaves via that tunnel and is released back into the cytosol. The cholesteryl ester then leaves via either the T tunnel into the membrane or through the L tunnel into the lumen of the cell. <ref name="Qian" /> | A main structural element of this enzyme is the tunnel systems. There are 3 main tunnels in each monomer: the cytosolic (C) tunnel opening to the cytosol, the transmembrane(T) tunnel opening to the membrane, and the lumenal(L) tunnel opens to the lumen. The C tunnel opens to the cytosol of the cell and is the entrance site for the Acyl CoA into the active site. Surface representations of SOAT indicate that there are 2 alpha helices that block the entrance to the C tunnel, therefore a conformational change needs to occur to move the 2 helices so the substrate can enter the tunnel. The T tunnel opens into the membrane and is where cholesterol enters to have access to the active site. The two substrates are catalyzed by the H460 in the active site to form the cholesteryl ester. The products then leave via different pathways. The CoA-SH in the C tunnel leaves via that tunnel and is released back into the cytosol. The cholesteryl ester then leaves via either the T tunnel into the membrane or through the L tunnel into the lumen of the cell. <ref name="Qian" /> | ||
| Line 23: | Line 23: | ||
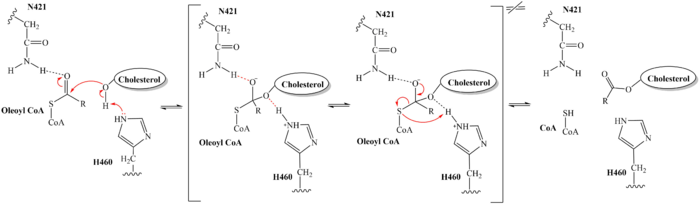
From the transition state, excess electron density on the carbonyl oxygen is collapsed back into a double bond. This causes the bond between the carbonyl carbon and sulfur to break, shifting electron density to the sulfur atom. To complete the mechanism, the negatively charged sulfur would reclaim the hydrogen from protonated H460. Acyl CoA would exit the active site as a [https://en.wikipedia.org/wiki/Leaving_group leaving group], leaving its R group attached to cholesterol in the form of a cholesterol ester. | From the transition state, excess electron density on the carbonyl oxygen is collapsed back into a double bond. This causes the bond between the carbonyl carbon and sulfur to break, shifting electron density to the sulfur atom. To complete the mechanism, the negatively charged sulfur would reclaim the hydrogen from protonated H460. Acyl CoA would exit the active site as a [https://en.wikipedia.org/wiki/Leaving_group leaving group], leaving its R group attached to cholesterol in the form of a cholesterol ester. | ||
| - | [[Image:6p2pMechanism.png|700 px|right|thumb|Figure 5. Esterification Reaction of SOAT with arrow pushing]] | + | [[Image:6p2pMechanism.png|700 px|right|thumb|'''Figure 5.''' Esterification Reaction of SOAT with arrow pushing]] |
It should be noted that this mechanism is largely hypothesized. Further analysis is needed to confirm the proposed steps. Additionally, mutations of W420A rendered the SOAT enzyme nonfunctional, indicating that it must be essential for catalytic activity. However, its role in the mechanism was not explicitly hypothesized. We believe that it plays a role in substrate binding through <scene name='87/879459/W420_intx/1'>hydrophobic interactions</scene> with CoenzymeA. | It should be noted that this mechanism is largely hypothesized. Further analysis is needed to confirm the proposed steps. Additionally, mutations of W420A rendered the SOAT enzyme nonfunctional, indicating that it must be essential for catalytic activity. However, its role in the mechanism was not explicitly hypothesized. We believe that it plays a role in substrate binding through <scene name='87/879459/W420_intx/1'>hydrophobic interactions</scene> with CoenzymeA. | ||
Revision as of 20:09, 24 April 2021
Human Sterol O-acyltransferase
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Guan C, Niu Y, Chen SC, Kang Y, Wu JX, Nishi K, Chang CCY, Chang TY, Luo T, Chen L. Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nat Commun. 2020 May 18;11(1):2478. doi: 10.1038/s41467-020-16288-4. PMID:32424158 doi:http://dx.doi.org/10.1038/s41467-020-16288-4
- ↑ 2.0 2.1 2.2 Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ 3.0 3.1 Bhattacharyya R, Kovacs DM. ACAT inhibition and amyloid beta reduction. Biochim Biophys Acta. 2010 Aug;1801(8):960-5. doi: 10.1016/j.bbalip.2010.04.003. , Epub 2010 Apr 14. PMID:20398792 doi:http://dx.doi.org/10.1016/j.bbalip.2010.04.003
- ↑ 4.0 4.1 Huttunen HJ, Kovacs DM. ACAT as a drug target for Alzheimer's disease. Neurodegener Dis. 2008;5(3-4):212-4. doi: 10.1159/000113705. Epub 2008 Mar 6. PMID:18322393 doi:http://dx.doi.org/10.1159/000113705
- ↑ Chang C, Dong R, Miyazaki A, Sakashita N, Zhang Y, Liu J, Guo M, Li BL, Chang TY. Human acyl-CoA:cholesterol acyltransferase (ACAT) and its potential as a target for pharmaceutical intervention against atherosclerosis. Acta Biochim Biophys Sin (Shanghai). 2006 Mar;38(3):151-6. doi:, 10.1111/j.1745-7270.2006.00154.x. PMID:16518538 doi:http://dx.doi.org/10.1111/j.1745-7270.2006.00154.x
- ↑ Ayyagari VN, Wang X, Diaz-Sylvester PL, Groesch K, Brard L. Assessment of acyl-CoA cholesterol acyltransferase (ACAT-1) role in ovarian cancer progression-An in vitro study. PLoS One. 2020 Jan 24;15(1):e0228024. doi: 10.1371/journal.pone.0228024., eCollection 2020. PMID:31978092 doi:http://dx.doi.org/10.1371/journal.pone.0228024
Student Contributors
- Kylie Pfeifer
- Stephanie Pellegrino
- Kaitlyn Roberts