We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Jacob Holt/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
| + | === Ligand Binding Pocket === | ||
| + | The ligand binding pocket is a narrow tunnel that extends approximately 24 Å into the mostly hydrophobic interior of the protein. The ligand is stabilized by bending into a kinked conformation which creates a tight fit in the binding pocket tunnel, and by a hydrogen bond that occurs between the W258 side chain and the acyl carbonyl<ref name="Bai" />. The kink in the tunnel is formed by the conserved residues, <scene name='87/877552/Desaturation_site/8'>T 257 and W 149</scene><ref name="Bai" />.7 which are stabilized by the hydrogen bond shared with Q143<ref name="Bai" />. There are <scene name='87/877552/Substrate_orientation_w_fe/5'>two Fe2+ ions</scene> that interact with the substrate; the Fe2+ ions are coordinated by 9 histidine residues. One metal ion is coordinated by 4 histidine's residues and a water molecule, and the other metal ion is coordinated by 5 histidine residues<ref name="Bai" />. <scene name='87/877552/Substrate_oreintation_fe_90deg/2'>When rotated 90 degrees</scene> the ligand is seen to be in a eclipsed position, indicating it is in its post-reaction form. The histidine residues position the metal ions 6.4 Å apart<ref name="Bai" />. | ||
| + | === Histidine Coordination === | ||
| + | There are <scene name='87/877552/Histidine_coordination/7'>9 invariant histidine residues</scene> that together coordinate the metal ions<ref name="Bai" />. One of the metal ions is coordinated by 4 histidine's residues and a water molecule, and the other metal ion is coordinated by 5 histidine residues<ref name="Bai" />. The histidine residues position the metal ions 6.4 Å apart<ref name="Bai" />. | ||
| - | + | === Desaturation Site === | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | === Desaturation | + | |
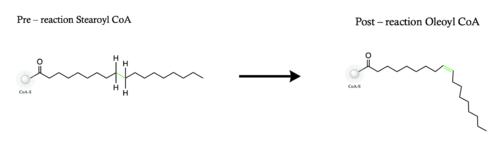
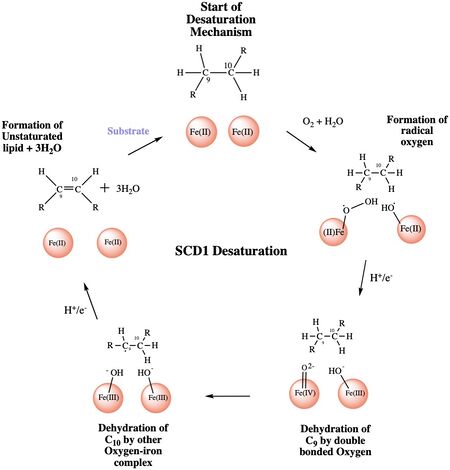
The ligand is desaturated at carbons 9 and 10<ref name="Shen" />. The desaturation site of the ligand takes place inside the active site tunnel which enforces correct positioning of the substrate<ref name="Bai" />. Before the reaction occurs, the ligand is in a gauche conformation at the desaturation site. This was determined by accidental usage of <scene name='87/877552/Pre_reaction_substrate_zn/2'>Zn+ ions</scene> which allowed for binding of the substrate but prevented the reaction<ref name="Shen" />. The product is in a cis conformation post-reaction. The product structure was determined using Fe2+ metal ions which allowed for the full reaction to take place<ref name="Shen" />. The difference between the <scene name='87/877552/Overlay_of_ligands/5'>substrate and product</scene> is the creation of a double bond, and the positioning of carbon 9 and 10 into a eclipsed position | The ligand is desaturated at carbons 9 and 10<ref name="Shen" />. The desaturation site of the ligand takes place inside the active site tunnel which enforces correct positioning of the substrate<ref name="Bai" />. Before the reaction occurs, the ligand is in a gauche conformation at the desaturation site. This was determined by accidental usage of <scene name='87/877552/Pre_reaction_substrate_zn/2'>Zn+ ions</scene> which allowed for binding of the substrate but prevented the reaction<ref name="Shen" />. The product is in a cis conformation post-reaction. The product structure was determined using Fe2+ metal ions which allowed for the full reaction to take place<ref name="Shen" />. The difference between the <scene name='87/877552/Overlay_of_ligands/5'>substrate and product</scene> is the creation of a double bond, and the positioning of carbon 9 and 10 into a eclipsed position | ||
| Line 111: | Line 82: | ||
== Biological Relevance == | == Biological Relevance == | ||
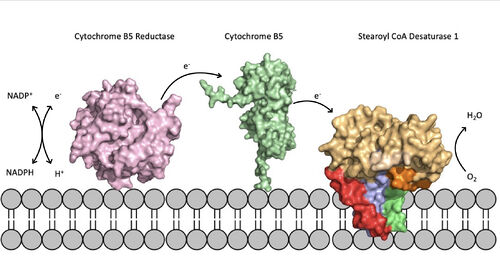
[[Image:scd_in_membrane.jpeg|500 px|thumb|Figure 5. Position of SCD within the biological membrane. It is part of an electron transport chain involving cytochrome b5 reductase and cytochrome b5 to allow for the activation of the catalytic molecule coordinated by the two ions in the center of SCD]]Absence or a deficit of SCD1 in the body is associated with obesity and insulin resistant which is a main cause of Type II diabetes [https://en.wikipedia.org/wiki/Type_2_diabetes type 2 diabetes]<ref name="Shen" />. Cancer sites in the body tend to show a much higher expression rate of SCD1<ref name="Shen" />. Focusing on SCD1 as a drug target could lead to advancements in treatment of obesity, diabetes, and other metabolic diseases<ref name="Bai" />. The ligand structure was determined by using Zn2+ metal ions and product structure was determined using Fe2+ ions<ref name="Shen" />. | [[Image:scd_in_membrane.jpeg|500 px|thumb|Figure 5. Position of SCD within the biological membrane. It is part of an electron transport chain involving cytochrome b5 reductase and cytochrome b5 to allow for the activation of the catalytic molecule coordinated by the two ions in the center of SCD]]Absence or a deficit of SCD1 in the body is associated with obesity and insulin resistant which is a main cause of Type II diabetes [https://en.wikipedia.org/wiki/Type_2_diabetes type 2 diabetes]<ref name="Shen" />. Cancer sites in the body tend to show a much higher expression rate of SCD1<ref name="Shen" />. Focusing on SCD1 as a drug target could lead to advancements in treatment of obesity, diabetes, and other metabolic diseases<ref name="Bai" />. The ligand structure was determined by using Zn2+ metal ions and product structure was determined using Fe2+ ions<ref name="Shen" />. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
== References == | == References == | ||
Revision as of 13:38, 25 April 2021
Desaturation of Fatty Acids using Stearoyl-CoA Desaturase-1 Enzyme
| |||||||||||
Student Contributions
Carson Maris, Jess Kersey, Jacob Holt