User:Megan Fleshman/Sandbox1
From Proteopedia
| Line 21: | Line 21: | ||
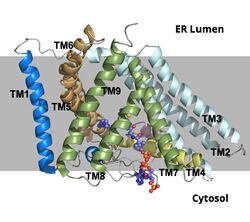
The active site is accessed through three tunnels that lead from the [http://en.wikipedia.org/wiki/Lumen_(anatomy) lumen], [https://en.wikipedia.org/wiki/Cytosol cytosol], and [https://en.wikipedia.org/wiki/Transmembrane_protein transmembrane] space to the center of the catalytic site. The tunnels allow the introduction of reactants into the acyl transferase mechanism and the exit of the products to the correct location depending on their function (figure 2). The cholesterol enters through the T tunnel while the acyl-CoA enters through the C tunnel. <ref name=”Qian”>PMID:32433614</ref> | The active site is accessed through three tunnels that lead from the [http://en.wikipedia.org/wiki/Lumen_(anatomy) lumen], [https://en.wikipedia.org/wiki/Cytosol cytosol], and [https://en.wikipedia.org/wiki/Transmembrane_protein transmembrane] space to the center of the catalytic site. The tunnels allow the introduction of reactants into the acyl transferase mechanism and the exit of the products to the correct location depending on their function (figure 2). The cholesterol enters through the T tunnel while the acyl-CoA enters through the C tunnel. <ref name=”Qian”>PMID:32433614</ref> | ||
The <scene name='87/877605/C_tunnel/4'>C tunnel</scene> is open to the cytosolic side of the protein in which the Acyl CoA enters into the catalytic domain. | The <scene name='87/877605/C_tunnel/4'>C tunnel</scene> is open to the cytosolic side of the protein in which the Acyl CoA enters into the catalytic domain. | ||
| - | The <scene name='87/877605/T_tunnel/2'>T tunnel</scene> is the transmembrane tunnel in which the cholesterol enters into the catalytic domain space. Important <scene name='87/877605/T_tunnel_residues/2'>residues</scene> of the T tunnel include Arg262, Phe263, and Leu306. These residues are important for the proper entrance and orientation of the cholesterol to allow for its deprotonation in the mechanism. Upon mutation of these residues, the tunnel function was inhibited. | + | The <scene name='87/877605/T_tunnel/2'>T tunnel</scene> is the transmembrane tunnel in which the cholesterol enters into the catalytic domain space. Important <scene name='87/877605/T_tunnel_residues/2'>residues</scene> of the T tunnel include Arg262, Phe263, and Leu306. These residues are important for the proper entrance and orientation of the cholesterol to allow for its deprotonation in the mechanism. Upon mutation of these residues, the tunnel function was inhibited. |
The <scene name='87/877605/L_tunnel/3'>L tunnel</scene> provides a potential opening to the lumen side. The enzymatic reaction occurs at the intersection of these two tunnels. It is catalyzed at the intersection of the two tunnels, where the His460 residue is located. The CoASH is released to the cytosol by way of the C tunnel, but the cholesterol ester either exits from the T tunnel to the membrane or through the L tunnel to the lumen. The specific mechanism by which the cholesterol ester product exits the tunnel to enter the lumen has not yet been determined. | The <scene name='87/877605/L_tunnel/3'>L tunnel</scene> provides a potential opening to the lumen side. The enzymatic reaction occurs at the intersection of these two tunnels. It is catalyzed at the intersection of the two tunnels, where the His460 residue is located. The CoASH is released to the cytosol by way of the C tunnel, but the cholesterol ester either exits from the T tunnel to the membrane or through the L tunnel to the lumen. The specific mechanism by which the cholesterol ester product exits the tunnel to enter the lumen has not yet been determined. | ||
Revision as of 21:30, 26 April 2021
Acyl-Coenzyme A: Cholesterol Acetyltransferase 1 (ACAT1): Function, Structure, and Inhibition
| |||||||||||
References
- ↑ Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ Farese RV Jr. The nine lives of ACAT inhibitors. Arterioscler Thromb Vasc Biol. 2006 Aug;26(8):1684-6. doi:, 10.1161/01.ATV.0000227511.35456.90. PMID:16857957 doi:http://dx.doi.org/10.1161/01.ATV.0000227511.35456.90
- ↑ Guan C, Niu Y, Chen SC, Kang Y, Wu JX, Nishi K, Chang CCY, Chang TY, Luo T, Chen L. Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nat Commun. 2020 May 18;11(1):2478. doi: 10.1038/s41467-020-16288-4. PMID:32424158 doi:http://dx.doi.org/10.1038/s41467-020-16288-4
- ↑ Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ Guan C, Niu Y, Chen SC, Kang Y, Wu JX, Nishi K, Chang CCY, Chang TY, Luo T, Chen L. Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nat Commun. 2020 May 18;11(1):2478. doi: 10.1038/s41467-020-16288-4. PMID:32424158 doi:http://dx.doi.org/10.1038/s41467-020-16288-4
- ↑ Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑
Mechanism
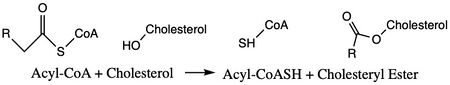
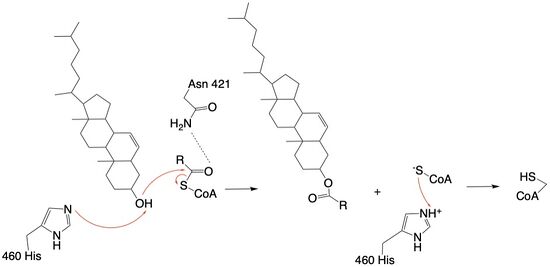
The mechanism of the [acyltransferace]reaction occurs in the catalytic site one of the monomers in the dimer of ACAT1. The T tunnel and and C tunnel converge to the same space to allow the proper orientation of the Acyl CoA and the incoming cholesterol from the transmembrane. The Acyl CoA is oriented in a way to allow the His460 to act as a base catalyst to begin the reaction by deprotonation of the cholesterol which allows it to attack the carbonyl carbon which breaks the sulfur carbonyl bond (figure 4). The H460 is positioned to deprotonate the cholesterol upon entering through the T tunnel: Acyl CoA upon entering is positioned to where the sulfur bonded to the carboxyl carbon is at the direct intersection of the T tunnel into the active site. The Acyl CoA is held in place by the of Asn 421. This mechanism produces Acyl-CoASH and cholesteryl ester. The Acyl-CoASH leaves through the C tunnel to the cytosol.
Inhibitor
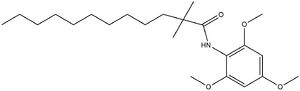
is known as a small molecule inhibitor that is part of the fatty acyl amide analog family, and functions as a competitive inhibitor of Acyl-CoA <ref>PMID:32424158</li> <li id="cite_note-Guan-8">↑ <sup>[[#cite_ref-Guan_8-0|9.0]]</sup> <sup>[[#cite_ref-Guan_8-1|9.1]]</sup> <sup>[[#cite_ref-Guan_8-2|9.2]]</sup> Guan C, Niu Y, Chen SC, Kang Y, Wu JX, Nishi K, Chang CCY, Chang TY, Luo T, Chen L. Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nat Commun. 2020 May 18;11(1):2478. doi: 10.1038/s41467-020-16288-4. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/32424158 32424158] doi:[http://dx.doi.org/10.1038/s41467-020-16288-4 http://dx.doi.org/10.1038/s41467-020-16288-4]</li> <li id="cite_note-Ayyagari-9">[[#cite_ref-Ayyagari_9-0|↑]] Ayyagari VN, Wang X, Diaz-Sylvester PL, Groesch K, Brard L. Assessment of acyl-CoA cholesterol acyltransferase (ACAT-1) role in ovarian cancer progression-An in vitro study. PLoS One. 2020 Jan 24;15(1):e0228024. doi: 10.1371/journal.pone.0228024., eCollection 2020. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/31978092 31978092] doi:[http://dx.doi.org/10.1371/journal.pone.0228024 http://dx.doi.org/10.1371/journal.pone.0228024]</li> <li id="cite_note-Vaziri-10">[[#cite_ref-Vaziri_10-0|↑]] Vaziri ND, Liang KH. Acyl-coenzyme A:cholesterol acyltransferase inhibition ameliorates proteinuria, hyperlipidemia, lecithin-cholesterol acyltransferase, SRB-1, and low-denisty lipoprotein receptor deficiencies in nephrotic syndrome. Circulation. 2004 Jul 27;110(4):419-25. doi: 10.1161/01.CIR.0000136023.70841.0F. , Epub 2004 Jul 19. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/15262831 15262831] doi:[http://dx.doi.org/10.1161/01.CIR.0000136023.70841.0F http://dx.doi.org/10.1161/01.CIR.0000136023.70841.0F]</li> <li id="cite_note-Willner-11">[[#cite_ref-Willner_11-0|↑]] Willner EL, Tow B, Buhman KK, Wilson M, Sanan DA, Rudel LL, Farese RV Jr. Deficiency of acyl CoA:cholesterol acyltransferase 2 prevents atherosclerosis in apolipoprotein E-deficient mice. Proc Natl Acad Sci U S A. 2003 Feb 4;100(3):1262-7. doi: 10.1073/pnas.0336398100., Epub 2003 Jan 21. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/12538880 12538880] doi:[http://dx.doi.org/10.1073/pnas.0336398100 http://dx.doi.org/10.1073/pnas.0336398100]</li> <li id="cite_note-name-12">↑ <sup>[[#cite_ref-name_12-0|13.0]]</sup> <sup>[[#cite_ref-name_12-1|13.1]]</sup> <sup>[[#cite_ref-name_12-2|13.2]]</sup> <sup>[[#cite_ref-name_12-3|13.3]]</sup> Chang TY, Chang CC, Bryleva E, Rogers MA, Murphy SR. Neuronal cholesterol esterification by ACAT1 in Alzheimer's disease. IUBMB Life. 2010 Apr;62(4):261-7. doi: 10.1002/iub.305. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/20101629 20101629] doi:[http://dx.doi.org/10.1002/iub.305 http://dx.doi.org/10.1002/iub.305]</li>
<li id="cite_note-Shibuya-13">↑ <sup>[[#cite_ref-Shibuya_13-0|14.0]]</sup> <sup>[[#cite_ref-Shibuya_13-1|14.1]]</sup> Shibuya Y, Chang CC, Chang TY. ACAT1/SOAT1 as a therapeutic target for Alzheimer's disease. Future Med Chem. 2015;7(18):2451-67. doi: 10.4155/fmc.15.161. Epub 2015 Dec 15. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/26669800 26669800] doi:[http://dx.doi.org/10.4155/fmc.15.161 http://dx.doi.org/10.4155/fmc.15.161]</li></ol></ref>
Student Contributors
- Megan Fleshman, Tori Templin, Haylie Moehlenkamp