User:Betsy Johns/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 29: | Line 29: | ||
=== Oleoyl-CoA Binding === | === Oleoyl-CoA Binding === | ||
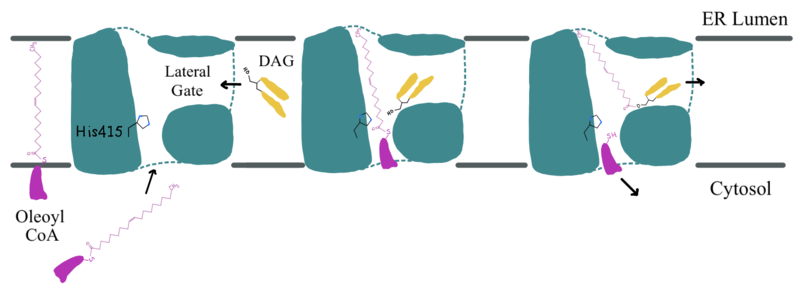
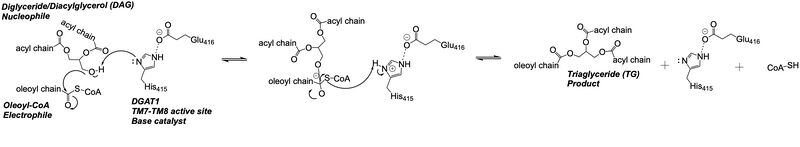
| - | Oleoyl-CoA enters DGAT1’s active site through a channel on the cytosolic side of the membrane. In order for the channel to widen and accommodate the fatty acid tail of the Oleoyl-CoA, <scene name='87/877512/Dgat_aligned2/17'>His415</scene> must flip from facing the cytosolic side to facing the lumenal side. It is speculated that the delta sulfur of the Met434 is involved in hydrogen bonding interactions with the catalytic His415 while it is flipped toward the cytosolic side of DGAT1. <ref name="Sui">PMID: 32433611</ref> The His415 needs to break these speculated hydrogen bonds with Met434 to flip toward the lumenal side of DGAT1. Once <scene name='87/877512/Acyl_coa/2'>Oleoyl-CoA</scene> is bound in the active site, residues Asn378, Gln437, Met434, and Gln465 stabilize the fatty acid tail within the cytosolic channel of the active site, while residues His415 and Gln416 are directly involved within the catalytic mechanism of DGAT1. <ref name="Sui">PMID: 32433611</ref> | + | Oleoyl-CoA enters DGAT1’s active site through a channel on the cytosolic side of the membrane. In order for the channel to widen and accommodate the fatty acid tail of the Oleoyl-CoA, <scene name='87/877512/Dgat_aligned2/17'>His415</scene> must flip from facing the cytosolic side to facing the lumenal side. It is speculated that the delta sulfur of the Met434 is involved in hydrogen bonding interactions with the catalytic His415 while it is flipped toward the cytosolic side of DGAT1 (6vyi). <ref name="Sui">PMID: 32433611</ref> The His415 needs to break these speculated hydrogen bonds with Met434 to flip toward the lumenal side of DGAT1. Once <scene name='87/877512/Acyl_coa/2'>Oleoyl-CoA</scene> is bound in the active site, residues Asn378, Gln437, Met434, and Gln465 stabilize the fatty acid tail within the cytosolic channel of the active site, while residues His415 and Gln416 are directly involved within the catalytic mechanism of DGAT1. <ref name="Sui">PMID: 32433611</ref> |
=== DAG Binding === | === DAG Binding === | ||
| - | <scene name='87/877512/Dag_binding/2'>DAG</scene> enters the active site through the lateral gate located in the lipid bilayer of the membrane. This lateral gate is a bent and hydrophobic channel that allows for hydrophobic linear or curvilinear molecules to enter <ref name="Sui">PMID: 32433611</ref> | + | <scene name='87/877512/Dag_binding/2'>DAG</scene> enters the active site through the lateral gate located in the lipid bilayer of the membrane. This lateral gate is a bent and hydrophobic channel that allows for hydrophobic linear or curvilinear molecules to enter. <ref name="Sui">PMID: 32433611</ref> The lateral gate channel is designed to allow for the entrance of DAG and the exit of a triacylglyceride. This channel is also lined with <scene name='87/877512/Lateral_gate_residues/4'>hydrophobic residues</scene> Phe342, Leu261, and Val381. <ref name="Wang">PMID: 32433610</ref><ref name="Sui">PMID: 32433611</ref> Once within the channel, DAG is positioned in close proximity to the bound Acyl-CoA and the catalytic His415. |
=== Active Site Structure === | === Active Site Structure === | ||
Revision as of 23:09, 26 April 2021
Diacylglycerol acyltransferase 1, DGAT1, synthesizes triacylglycerides
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Wang L, Qian H, Nian Y, Han Y, Ren Z, Zhang H, Hu L, Prasad BVV, Laganowsky A, Yan N, Zhou M. Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature. 2020 May;581(7808):329-332. doi: 10.1038/s41586-020-2280-2. Epub 2020 May, 13. PMID:32433610 doi:http://dx.doi.org/10.1038/s41586-020-2280-2
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 Sui X, Wang K, Gluchowski NL, Elliott SD, Liao M, Walther TC, Farese RV Jr. Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature. 2020 May;581(7808):323-328. doi: 10.1038/s41586-020-2289-6. Epub 2020 May, 13. PMID:32433611 doi:http://dx.doi.org/10.1038/s41586-020-2289-6
- ↑ 3.0 3.1 3.2 Ma D, Wang Z, Merrikh CN, Lang KS, Lu P, Li X, Merrikh H, Rao Z, Xu W. Crystal structure of a membrane-bound O-acyltransferase. Nature. 2018 Oct;562(7726):286-290. doi: 10.1038/s41586-018-0568-2. Epub 2018 Oct, 3. PMID:30283133 doi:http://dx.doi.org/10.1038/s41586-018-0568-2
- ↑ 4.0 4.1 4.2 4.3 Denison H, Nilsson C, Lofgren L, Himmelmann A, Martensson G, Knutsson M, Al-Shurbaji A, Tornqvist H, Eriksson JW. Diacylglycerol acyltransferase 1 inhibition with AZD7687 alters lipid handling and hormone secretion in the gut with intolerable side effects: a randomized clinical trial. Diabetes Obes Metab. 2014 Apr;16(4):334-43. doi: 10.1111/dom.12221. Epub 2013 Oct, 31. PMID:24118885 doi:http://dx.doi.org/10.1111/dom.12221
- ↑ Stephen J, Vilboux T, Haberman Y, Pri-Chen H, Pode-Shakked B, Mazaheri S, Marek-Yagel D, Barel O, Di Segni A, Eyal E, Hout-Siloni G, Lahad A, Shalem T, Rechavi G, Malicdan MC, Weiss B, Gahl WA, Anikster Y. Congenital protein losing enteropathy: an inborn error of lipid metabolism due to DGAT1 mutations. Eur J Hum Genet. 2016 Aug;24(9):1268-73. doi: 10.1038/ejhg.2016.5. Epub 2016 Feb , 17. PMID:26883093 doi:http://dx.doi.org/10.1038/ejhg.2016.5
- ↑ Villanueva CJ, Monetti M, Shih M, Zhou P, Watkins SM, Bhanot S, Farese RV Jr. Specific role for acyl CoA:Diacylglycerol acyltransferase 1 (Dgat1) in hepatic steatosis due to exogenous fatty acids. Hepatology. 2009 Aug;50(2):434-42. doi: 10.1002/hep.22980. PMID:19472314 doi:http://dx.doi.org/10.1002/hep.22980
Student Contributors
- Betsy Johns
- Elise Wang
- Tyler Bihasa