This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Jacob Holt/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 44: | Line 44: | ||
=== Ligand Binding Pocket and Histidine Coordination === | === Ligand Binding Pocket and Histidine Coordination === | ||
| - | The ligand binding pocket is a narrow tunnel that extends approximately 24 Å into the mostly hydrophobic interior of the protein. The ligand is stabilized by bending into a kinked conformation which creates a tight fit in the binding pocket tunnel, and by a hydrogen bond that occurs between the <scene name='87/877552/W258/2'>W 258</scene> side chain and the acyl carbonyl<ref name="Bai" />. The kink in the tunnel is formed by the conserved residues, <scene name='87/877552/Desaturation_site/9'>T 257 and W 149</scene> which are stabilized by the hydrogen bond shared with Q143<ref name="Bai" />. There are <scene name='87/877552/Substrate_orientation_w_fe/5'>two Fe2+ ions</scene> that interact with the substrate; the Fe2+ ions are coordinated by 9 histidine residues. <scene name='87/877552/Substrate_oreintation_fe_90deg/3'>When rotated 90 degrees</scene> the ligand is seen to be in a eclipsed position, indicating it is in its post-reaction form. One metal ion is coordinated by 4 histidines residues and a water molecule, and the other metal ion is coordinated by 5 histidine residues<ref name="Bai" />. The histidine residues position the metal ions 6.4 Å apart<ref name="Bai" />. | + | The ligand binding pocket is a narrow tunnel that extends approximately 24 Å into the mostly hydrophobic interior of the protein. The ligand is stabilized by bending into a kinked conformation which creates a tight fit in the binding pocket tunnel, and by a hydrogen bond that occurs between the <scene name='87/877552/W258/2'>W 258</scene> side chain and the acyl carbonyl<ref name="Bai" />. The kink in the tunnel is formed by the conserved residues, <scene name='87/877552/Desaturation_site/9'>T 257 and W 149</scene> which are stabilized by the hydrogen bond shared with Q143<ref name="Bai" />. There are <scene name='87/877552/Substrate_orientation_w_fe/5'>two Fe2+ ions</scene> that interact with the substrate; the Fe2+ ions are coordinated by <scene name='87/877552/Histidine_coordination/8'>9 invariant histidine residues</scene>. <scene name='87/877552/Substrate_oreintation_fe_90deg/3'>When rotated 90 degrees</scene> the ligand is seen to be in a eclipsed position, indicating it is in its post-reaction form. One metal ion is coordinated by 4 histidines residues and a water molecule, and the other metal ion is coordinated by 5 histidine residues<ref name="Bai" />. The histidine residues position the metal ions 6.4 Å apart<ref name="Bai" />. |
=== Desaturation Site === | === Desaturation Site === | ||
| Line 53: | Line 53: | ||
=== Catalytic Molecule === | === Catalytic Molecule === | ||
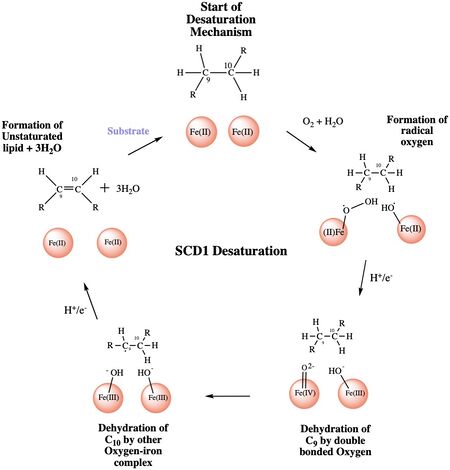
| - | The <scene name='87/877552/Water/3'>catalytic molecule</scene> of the SCD1 enzyme is a water molecule, coordinated by <scene name='87/877552/Asparagine_h20_stabilization/ | + | The <scene name='87/877552/Water/3'>catalytic molecule</scene> of the SCD1 enzyme is a water molecule, coordinated by <scene name='87/877552/Asparagine_h20_stabilization/2'>N 261</scene> via hydrogen bonding<ref name="Bai" />. The water molecule is 2.2 Å away from the Fe2+ metal ion molecule<ref name="Bai" />. It interacts with the Fe2+ ion to make highly reactive radicals that are able to desaturate the highly stable carbon chain<ref name="Yu">DOI:10.1021/acscatal.9b00456 </ref>. It is through the coordination of these ions by the histidines that the <scene name='87/877552/Diiron_center/8'>substrate and catalytic molecule</scene> are able to be positioned within the vicinity of carbons 9 and 10 of the ligand<ref name="Yu" />. |
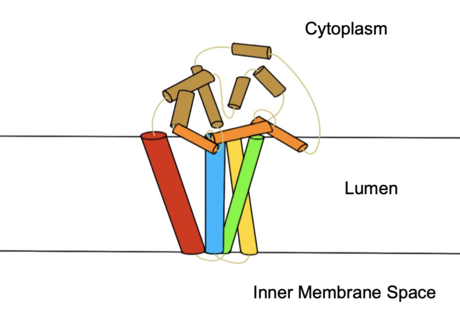
=== Substrate Entering and Leaving === | === Substrate Entering and Leaving === | ||
| Line 93: | Line 93: | ||
The SCD1 enzyme can be down regulated or slightly inhibited by low-carbohydrate diets, low glucose levels, low insulin levels, and a low cholesterol diet. SCD1 is also regulated by leptin [https://en.wikipedia.org/wiki/Leptin leptin]; a protein that is created by fat cells<ref name="Ntambi" />. When the leptin signaling pathway is fully functioning, the SCD1 enzyme is regulated in its ability to create MUFAs. SCD1 being associated with obesity could directly be tied to errors in the signaling pathways of the leptin protein<ref name="Ntambi" />. | The SCD1 enzyme can be down regulated or slightly inhibited by low-carbohydrate diets, low glucose levels, low insulin levels, and a low cholesterol diet. SCD1 is also regulated by leptin [https://en.wikipedia.org/wiki/Leptin leptin]; a protein that is created by fat cells<ref name="Ntambi" />. When the leptin signaling pathway is fully functioning, the SCD1 enzyme is regulated in its ability to create MUFAs. SCD1 being associated with obesity could directly be tied to errors in the signaling pathways of the leptin protein<ref name="Ntambi" />. | ||
| - | == Regulation | + | == Regulation == |
| - | The main function of SCD1 is to create the desaturated ligand that is used in the synthesis of cholesterol esters and triglycerides<ref name="Shen" />. Based on this main function it has been confirmed that insulin and carbohydrate metabolism play a major role in the regulation of SCD1<ref name="Ntambi">PMID: 7480063</ref>. Along with insulin many other hormones have shown positive regulation of the SCD1 enzyme including T3, estradiol, and dexamethasone<ref name="Ntambi" />. The carbohydrate metabolism can be altered by eating a fat-free, high carbohydrate diet with an increased intake of polyunsaturated fatty acids<ref name="Ntambi" />. The will negatively regulate the mRNA expression of the SCD1 enzyme in the liver because high amounts of carbohydrates and polyunsaturated fatty acids decrease/inhibit the activity of the SCD1 enzyme<ref name="Ntambi" />. | + | The main function of SCD1 is to create the desaturated ligand that is used in the synthesis of cholesterol esters and triglycerides<ref name="Shen" />. Based on this main function it has been confirmed that insulin and carbohydrate metabolism play a major role in the regulation of SCD1<ref name="Ntambi">PMID: 7480063</ref>. Along with insulin many other hormones have shown positive regulation of the SCD1 enzyme including [https://en.wikipedia.org/wiki/Triiodothyronine triiodothyronine] (T3), [https://en.wikipedia.org/wiki/Estradiol estradiol], and [https://en.wikipedia.org/wiki/Dexamethasone dexamethasone]<ref name="Ntambi" />. The carbohydrate metabolism can be altered by eating a fat-free, high carbohydrate diet with an increased intake of polyunsaturated fatty acids<ref name="Ntambi" />. The will negatively regulate the mRNA expression of the SCD1 enzyme in the liver because high amounts of carbohydrates and polyunsaturated fatty acids decrease/inhibit the activity of the SCD1 enzyme<ref name="Ntambi" />. |
== References == | == References == | ||
Revision as of 23:27, 26 April 2021
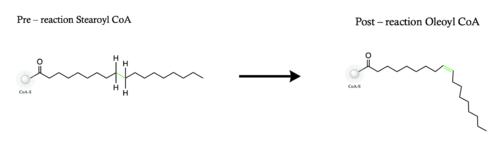
Desaturation of Fatty Acids using Stearoyl-CoA Desaturase-1 Enzyme
| |||||||||||
Student Contributions
Carson Maris, Jess Kersey, Jacob Holt