We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox GGC3
From Proteopedia
(Difference between revisions)
| Line 41: | Line 41: | ||
== Structural highlights == | == Structural highlights == | ||
| - | The conserved catalytic lysine for the adenylation reaction<ref name="conserved">Branchini, B. R., Murtiashaw, M. H., Magyar, R. A., Anderson, S. M. (2000). The Role of Lysine 529, a Conserved Residue of the Acyl-Adenylate-Forming Enzyme Superfamily, in Firefly Luciferase. ''Biochemistry 39''(18), 5433-5440. https://doi.org/10.1021/bi9928804</ref>, Lys529, interacts with the carbonyl oxygen of the adenylate, the O5 atom that bridges the ribose and sulfamate moiety, and the main chain carbonyl of Gly316. The second conformation observations show that the side chain amine of Lys443 adopts a nearly identical position as Lys529, and Gln448 of the C-terminal domain rotates into the binding pocket where it interacts with a sulfamate oxygen<ref name="Sundlov"/>. Altogether (with the inclusion of an ionic interaction between Glu479 and Arg437), these interactions are responsible for the stabilization of the new C-terminal conformation. | + | The conserved catalytic lysine for the adenylation reaction<ref name="conserved">Branchini, B. R., Murtiashaw, M. H., Magyar, R. A., Anderson, S. M. (2000). The Role of Lysine 529, a Conserved Residue of the Acyl-Adenylate-Forming Enzyme Superfamily, in Firefly Luciferase. ''Biochemistry 39''(18), 5433-5440. https://doi.org/10.1021/bi9928804</ref>, Lys529, interacts with the carbonyl oxygen of the adenylate, the O5 atom that bridges the ribose and sulfamate moiety, and the main chain carbonyl of Gly316. The second conformation observations show that the side chain amine of Lys443 adopts a nearly identical position as Lys529, and Gln448 of the C-terminal domain rotates into the binding pocket where it interacts with a sulfamate oxygen<ref name="Sundlov"/><ref name="Bruce">Branchini, B. R., Southworth, T. L., Murtiahsaw, M. H., Wilkinson, S. R., Khattak, N. F., Rosenberg, J. C., & Zimmer, M. (2005). Mutagenesis Evidence that the Partial Reactions of Firefly Bioluminescence are Catalyzed by Different Conformations of the Luciferase C-Terminal Domain. “Biochemistry 44”(5), 1385-1393. https://doi.org/10.1021/bi047903f</ref>. Altogether (with the inclusion of an ionic interaction between Glu479 and Arg437), these interactions are responsible for the stabilization of the new C-terminal conformation. |
(going through changes but math is cool :/)The A8 motif harbors the hinge residue at Lys439 and the antiparallel two stranded β-sheet is directed into the active site of the enzyme. The φ/ψ angles of Lys439 change from −73°/−12° in the structure of wild-type luciferase in the adenylate-forming conformation to −69°/158° in the cross- linked structure.this illustrates that a large component of the conformational change occurs with a rotation of the ψ angle of the hinge residue. Additional torsion angle changes are seen in φ angles for Arg437 and Leu441, although the magnitude of the change is not as large as at the hinge residue Lys439<ref name="Sundlov"/>. | (going through changes but math is cool :/)The A8 motif harbors the hinge residue at Lys439 and the antiparallel two stranded β-sheet is directed into the active site of the enzyme. The φ/ψ angles of Lys439 change from −73°/−12° in the structure of wild-type luciferase in the adenylate-forming conformation to −69°/158° in the cross- linked structure.this illustrates that a large component of the conformational change occurs with a rotation of the ψ angle of the hinge residue. Additional torsion angle changes are seen in φ angles for Arg437 and Leu441, although the magnitude of the change is not as large as at the hinge residue Lys439<ref name="Sundlov"/>. | ||
Revision as of 15:43, 27 April 2021
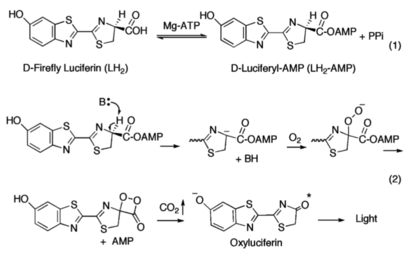
Firefly Luciferase
tttaaarrgggetttt to the right plaaccceee and finishh :(
| |||||||||||
References
- ↑ Branchini, B. R., Magyar, R. A., Murtiashaw, M. H., Anderson, S. M., Helgerson, L. C., & Zimmer, M. (1999). Site-directed mutagenesis of firefly luciferase active site amino acids: a proposed model for bioluminescence color. Biochemistry 38(40), 13223–13230. https://doi.org/10.1021/bi991181o
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Sundlov, J. A., Fontaine, D. M., Southworth, T. L., Branchini, B. R., Gulick, A. M. (2012). Crystal Structure of Firefly Luciferase in a Second Catalytic Conformation Supports a Domain Alternation Mechanism. Biochemistry 51(33), 6493-6495. https://doi.org/10.1021/bi300934s
- ↑ Marahiel, M. A., Stachelhaus, T., Mootz, H. D. (1997). Modular Peptide Synthetases Involved in Nonribosmal Peptide Synthesis. Chemical Reviews 97(7), 2651-2674. https://doi.org/10.1021/cr960029e
- ↑ Branchini, B. R., Murtiashaw, M. H., Magyar, R. A., Anderson, S. M. (2000). The Role of Lysine 529, a Conserved Residue of the Acyl-Adenylate-Forming Enzyme Superfamily, in Firefly Luciferase. Biochemistry 39(18), 5433-5440. https://doi.org/10.1021/bi9928804
- ↑ Branchini, B. R., Southworth, T. L., Murtiahsaw, M. H., Wilkinson, S. R., Khattak, N. F., Rosenberg, J. C., & Zimmer, M. (2005). Mutagenesis Evidence that the Partial Reactions of Firefly Bioluminescence are Catalyzed by Different Conformations of the Luciferase C-Terminal Domain. “Biochemistry 44”(5), 1385-1393. https://doi.org/10.1021/bi047903f