We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Megan Fleshman/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
==Structural Overview== | ==Structural Overview== | ||
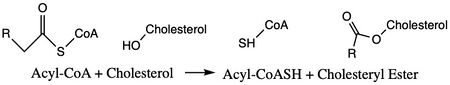
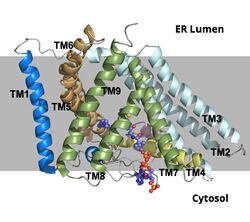
| - | ACAT1 was experimentally determined as a <scene name='87/877604/Tetramer/4'>tetramer</scene> of size approximately 260 kDa, composed of helices and loops <ref name= | + | ACAT1 was experimentally determined as a <scene name='87/877604/Tetramer/4'>tetramer</scene> of size approximately 260 kDa, composed of helices and loops <ref name="Guan">PMID:32424158</ref>. Each monomer of the [http://en.wikipedia.org/wiki/Tetramer tetramer] contains 9 transmembrane helices (Figure 3). However, the <scene name='87/877605/Colored_dimer/2'>dimer of ACAT1</scene> was found to be the biologically active arrangement. |
[[Image:Screen_Shot_2021-04-20_at_3.29.54_PM.jpg|400 px|left|thumb|Figure 2. ACAT1 Dimer in the Membrane. The gray shaded region is the plane of the bilipid membrane. The tunnels where molecules enter and exit are labeled. The ER lumen is at the top with the cytosol at the bottom.]] | [[Image:Screen_Shot_2021-04-20_at_3.29.54_PM.jpg|400 px|left|thumb|Figure 2. ACAT1 Dimer in the Membrane. The gray shaded region is the plane of the bilipid membrane. The tunnels where molecules enter and exit are labeled. The ER lumen is at the top with the cytosol at the bottom.]] | ||
===Dimer-Dimer Interface=== | ===Dimer-Dimer Interface=== | ||
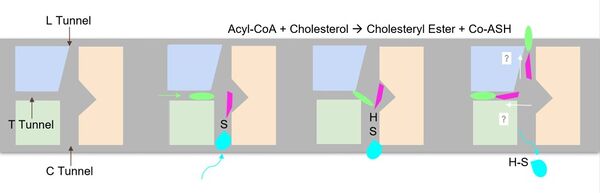
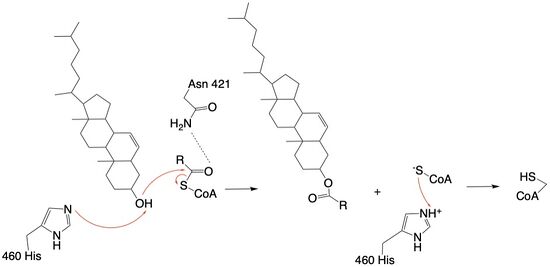
| - | The <scene name='87/877604/Dimer_interface/19'>dimer-dimer interface</scene> is mobile and mostly hydrophobic, and the residues interact in a shape-complementary manner <ref name= | + | The <scene name='87/877604/Dimer_interface/19'>dimer-dimer interface</scene> is mobile and mostly hydrophobic, and the residues interact in a shape-complementary manner <ref name="Guan">PMID:32424158</ref>. It was also found that the reaction chamber is shielded by a lid from the cytosolic side, which leads to low catalytic activity. The binding of Acyl-CoA and cholesterol induce conformational changes that activate the tunnels necessary for substrates to enter them. Work is still being done to fully determine the mechanism of this reaction, but this is the proposed pathway <ref name=”Qian”>PMID:32433614</ref>. [[Image:Screen_Shot_2021-04-20_at_3.30.39_PM.jpg|250 px|right|thumb|Figure 3. ACAT1 Monomer in the Membrane. This shows the 9 transmembrane helices. Each helix is labeled and colored according to the different domains. The ligand is shown as ball and stick.]] |
===Tunnels=== | ===Tunnels=== | ||
Revision as of 19:07, 27 April 2021
Acyl-Coenzyme A: Cholesterol Acetyltransferase 1 (ACAT1): Function, Structure, and Inhibition
| |||||||||||
References
- ↑ Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ Farese RV Jr. The nine lives of ACAT inhibitors. Arterioscler Thromb Vasc Biol. 2006 Aug;26(8):1684-6. doi:, 10.1161/01.ATV.0000227511.35456.90. PMID:16857957 doi:http://dx.doi.org/10.1161/01.ATV.0000227511.35456.90
- ↑ 3.0 3.1 3.2 3.3 3.4 Guan C, Niu Y, Chen SC, Kang Y, Wu JX, Nishi K, Chang CCY, Chang TY, Luo T, Chen L. Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nat Commun. 2020 May 18;11(1):2478. doi: 10.1038/s41467-020-16288-4. PMID:32424158 doi:http://dx.doi.org/10.1038/s41467-020-16288-4
- ↑ Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ 9.0 9.1 9.2 9.3 9.4 Guan C, Niu Y, Chen SC, Kang Y, Wu JX, Nishi K, Chang CCY, Chang TY, Luo T, Chen L. Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nat Commun. 2020 May 18;11(1):2478. doi: 10.1038/s41467-020-16288-4. PMID:32424158 doi:http://dx.doi.org/10.1038/s41467-020-16288-4
- ↑ Ayyagari VN, Wang X, Diaz-Sylvester PL, Groesch K, Brard L. Assessment of acyl-CoA cholesterol acyltransferase (ACAT-1) role in ovarian cancer progression-An in vitro study. PLoS One. 2020 Jan 24;15(1):e0228024. doi: 10.1371/journal.pone.0228024., eCollection 2020. PMID:31978092 doi:http://dx.doi.org/10.1371/journal.pone.0228024
- ↑ Vaziri ND, Liang KH. Acyl-coenzyme A:cholesterol acyltransferase inhibition ameliorates proteinuria, hyperlipidemia, lecithin-cholesterol acyltransferase, SRB-1, and low-denisty lipoprotein receptor deficiencies in nephrotic syndrome. Circulation. 2004 Jul 27;110(4):419-25. doi: 10.1161/01.CIR.0000136023.70841.0F. , Epub 2004 Jul 19. PMID:15262831 doi:http://dx.doi.org/10.1161/01.CIR.0000136023.70841.0F
- ↑ Willner EL, Tow B, Buhman KK, Wilson M, Sanan DA, Rudel LL, Farese RV Jr. Deficiency of acyl CoA:cholesterol acyltransferase 2 prevents atherosclerosis in apolipoprotein E-deficient mice. Proc Natl Acad Sci U S A. 2003 Feb 4;100(3):1262-7. doi: 10.1073/pnas.0336398100., Epub 2003 Jan 21. PMID:12538880 doi:http://dx.doi.org/10.1073/pnas.0336398100
- ↑ 13.0 13.1 Shibuya Y, Chang CC, Chang TY. ACAT1/SOAT1 as a therapeutic target for Alzheimer's disease. Future Med Chem. 2015;7(18):2451-67. doi: 10.4155/fmc.15.161. Epub 2015 Dec 15. PMID:26669800 doi:http://dx.doi.org/10.4155/fmc.15.161
Student Contributors
- Megan Fleshman, Tori Templin, Haylie Moehlenkamp