We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox GGC3
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
==Firefly Luciferase== | ==Firefly Luciferase== | ||
| - | + | apple juice! B~) | |
| - | + | ||
<StructureSection loadfiles='4G36''4G37' size='340' side='right' caption='Luciferin-4-monooxygenase. The wild-type luciferase in the adenylate-forming conformation with DLSA (PDB 4G36) and the cross-linked luciferase in the second catalytic conformation with DLSA (PDB 4G37)' scene=''> | <StructureSection loadfiles='4G36''4G37' size='340' side='right' caption='Luciferin-4-monooxygenase. The wild-type luciferase in the adenylate-forming conformation with DLSA (PDB 4G36) and the cross-linked luciferase in the second catalytic conformation with DLSA (PDB 4G37)' scene=''> | ||
Firefly luciferase, of the common eastern firefly (''Photinus pyralis''), is responsible for the ability of the firefly to exhibit bioluminescence. The enzyme luciferin-4-monoxygenase, which catalyzes a multistep oxidative decarboxylation of the luciferyl-AMP intermediate (LH<sub>2</sub>-AMP) to produce bioluminescence, is a part of the ANL superfamily named so after the '''a'''cyl-CoA syntheses, the adenylation domains of the modular '''n'''on-ribosomal peptide synthetases (NRPs), and '''l'''uciferase. | Firefly luciferase, of the common eastern firefly (''Photinus pyralis''), is responsible for the ability of the firefly to exhibit bioluminescence. The enzyme luciferin-4-monoxygenase, which catalyzes a multistep oxidative decarboxylation of the luciferyl-AMP intermediate (LH<sub>2</sub>-AMP) to produce bioluminescence, is a part of the ANL superfamily named so after the '''a'''cyl-CoA syntheses, the adenylation domains of the modular '''n'''on-ribosomal peptide synthetases (NRPs), and '''l'''uciferase. | ||
| Line 16: | Line 15: | ||
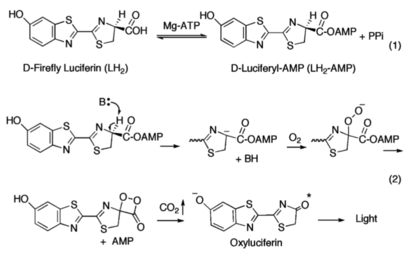
[[Image:Mechanism_of_Firefly_Bioluminescence.png|thumb|upright=2.3|The generally accepted mechanism of firefly bioluminescence. The first reaction involves the production of an luciferyl-adenylate intermediate (1). The second reaction involves oxidative decarboxylation that emits CO<sub>2</sub> and results in bioluminescent properties(2).<ref name="Sundlov"/>]] | [[Image:Mechanism_of_Firefly_Bioluminescence.png|thumb|upright=2.3|The generally accepted mechanism of firefly bioluminescence. The first reaction involves the production of an luciferyl-adenylate intermediate (1). The second reaction involves oxidative decarboxylation that emits CO<sub>2</sub> and results in bioluminescent properties(2).<ref name="Sundlov"/>]] | ||
Nothing but pain. Hi! Im Paul<ref name="Sundlov"/><ref name="Bruce">Branchini, B. R., Southworth, T. L., Murtiahsaw, M. H., Wilkinson, S. R., Khattak, N. F., Rosenberg, J. C., & Zimmer, M. (2005). Mutagenesis Evidence that the Partial Reactions of Firefly Bioluminescence are Catalyzed by Different Conformations of the Luciferase C-Terminal Domain. “Biochemistry 44”(5), 1385-1393. https://doi.org/10.1021/bi047903f</ref><ref name="Oba">Oba, Y., Ojika, M., Inouye, S. (2003). Firefly luciferase is a bifunctional enzyme: ATP-dependent monoxygenase and a long chain fatty acyl-CoA synthetase. “FEBS Letters 540”(1-3), 251-254. https://doi.org/10.1016/S0014-5793(03)00272-2</ref><ref name="Nakamura">Nakamura, M., Maki, S., Amano, Y., Ohkita, Y., Niwa, K., Hirano, T., Ohmiya, Y., & Niwa, H. (2005). Firefly luciferase exhibits bimodal action depending on the luciferin chirality. “Biochemical and Biophysical Research Communications, 331”(2), 471–475. https://doi.org/10.1016/j.bbrc.2005.03.202</ref>. | Nothing but pain. Hi! Im Paul<ref name="Sundlov"/><ref name="Bruce">Branchini, B. R., Southworth, T. L., Murtiahsaw, M. H., Wilkinson, S. R., Khattak, N. F., Rosenberg, J. C., & Zimmer, M. (2005). Mutagenesis Evidence that the Partial Reactions of Firefly Bioluminescence are Catalyzed by Different Conformations of the Luciferase C-Terminal Domain. “Biochemistry 44”(5), 1385-1393. https://doi.org/10.1021/bi047903f</ref><ref name="Oba">Oba, Y., Ojika, M., Inouye, S. (2003). Firefly luciferase is a bifunctional enzyme: ATP-dependent monoxygenase and a long chain fatty acyl-CoA synthetase. “FEBS Letters 540”(1-3), 251-254. https://doi.org/10.1016/S0014-5793(03)00272-2</ref><ref name="Nakamura">Nakamura, M., Maki, S., Amano, Y., Ohkita, Y., Niwa, K., Hirano, T., Ohmiya, Y., & Niwa, H. (2005). Firefly luciferase exhibits bimodal action depending on the luciferin chirality. “Biochemical and Biophysical Research Communications, 331”(2), 471–475. https://doi.org/10.1016/j.bbrc.2005.03.202</ref>. | ||
| + | |||
| Line 41: | Line 41: | ||
== Structural highlights == | == Structural highlights == | ||
| + | [[Image:Common_Eastern_Firefly.jpg|thumb|right|upright=1.7|The φ/ψ angles of the Lys439 residue undergo a rotational transformation of −73°/−12° to −69°/158° in the adenylate-forming to second-catalytic conformation, respectively, showing that the major torsional change in the ψ angle is observed<ref name="Sundlov"/>.]] | ||
The conserved catalytic lysine for the adenylation reaction<ref name="conserved">Branchini, B. R., Murtiashaw, M. H., Magyar, R. A., Anderson, S. M. (2000). The Role of Lysine 529, a Conserved Residue of the Acyl-Adenylate-Forming Enzyme Superfamily, in Firefly Luciferase. ''Biochemistry 39''(18), 5433-5440. https://doi.org/10.1021/bi9928804</ref>, Lys529, interacts with the carbonyl oxygen of the adenylate, the O5 atom that bridges the ribose and sulfamate moiety, and the main chain carbonyl of Gly316. The second conformation observations show that the side chain amine of Lys443 adopts a nearly identical position as Lys529, and Gln448 of the C-terminal domain rotates into the binding pocket where it interacts with a sulfamate oxygen<ref name="Sundlov"/><ref name="Bruce"/>. Altogether (with the inclusion of an ionic interaction between Glu479 and Arg437), these interactions are responsible for the stabilization of the new C-terminal conformation. | The conserved catalytic lysine for the adenylation reaction<ref name="conserved">Branchini, B. R., Murtiashaw, M. H., Magyar, R. A., Anderson, S. M. (2000). The Role of Lysine 529, a Conserved Residue of the Acyl-Adenylate-Forming Enzyme Superfamily, in Firefly Luciferase. ''Biochemistry 39''(18), 5433-5440. https://doi.org/10.1021/bi9928804</ref>, Lys529, interacts with the carbonyl oxygen of the adenylate, the O5 atom that bridges the ribose and sulfamate moiety, and the main chain carbonyl of Gly316. The second conformation observations show that the side chain amine of Lys443 adopts a nearly identical position as Lys529, and Gln448 of the C-terminal domain rotates into the binding pocket where it interacts with a sulfamate oxygen<ref name="Sundlov"/><ref name="Bruce"/>. Altogether (with the inclusion of an ionic interaction between Glu479 and Arg437), these interactions are responsible for the stabilization of the new C-terminal conformation. | ||
| - | + | I...maybe will write more. maybe. | |
| - | + | ||
| - | + | ||
== Relevance == | == Relevance == | ||
| - | [[Image:Photinus_pyralis.jpg|thumb|The Common Eastern Firefly expressing bioluminescence seen giving off a yellow-green hue.]] | + | [[Image:Photinus_pyralis.jpg|thumb|left|The Common Eastern Firefly expressing bioluminescence seen giving off a yellow-green hue.]] |
Firefly luciferase has successfully been shown to act as modulatory bioluminescent indicator in the detection and quantification of protein kinase A activation in living cells <ref name="Newby">Sala-Newby, G. B., & Campbell, A. K. (1991). Engineering a bioluminescent indicator for cyclic AMP-dependent protein kinase. “The Biochemical Journal”, 279 (Pt 3), 727–732. https://doi.org/10.1042/bj2790727</ref>. Further, due to its bioluminescent sensitivity, firefly luciferase has been utilized in assays as a genetic reporter in eukaryotic cells<ref name="de">de Wet, J. R., Wood, K. V., DeLuca, M., Helinski, D. R., & Subramani, S. (1987). Firefly luciferase gene: structure and expression in mammalian cells. Molecular and cellular biology, 7(2), 725–737. https://doi.org/10.1128/mcb.7.2.725</ref><ref name="Escherichia">de Wet, J. R., Wood, K. V., Helinski, D. R., & DeLuca, M. (1985). Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America, 82(23), 7870–7873. https://doi.org/10.1073/pnas.82.23.7870</ref><ref name="Throne">Thorne, N., Shen, M., Lea, W. A., Simeonov, A., Lovell, S., Auld, D. S., & Inglese, J. (2012). Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter. Chemistry & biology, 19(8), 1060–1072. https://doi.org/10.1016/j.chembiol.2012.07.015</ref>, amongst other things. | Firefly luciferase has successfully been shown to act as modulatory bioluminescent indicator in the detection and quantification of protein kinase A activation in living cells <ref name="Newby">Sala-Newby, G. B., & Campbell, A. K. (1991). Engineering a bioluminescent indicator for cyclic AMP-dependent protein kinase. “The Biochemical Journal”, 279 (Pt 3), 727–732. https://doi.org/10.1042/bj2790727</ref>. Further, due to its bioluminescent sensitivity, firefly luciferase has been utilized in assays as a genetic reporter in eukaryotic cells<ref name="de">de Wet, J. R., Wood, K. V., DeLuca, M., Helinski, D. R., & Subramani, S. (1987). Firefly luciferase gene: structure and expression in mammalian cells. Molecular and cellular biology, 7(2), 725–737. https://doi.org/10.1128/mcb.7.2.725</ref><ref name="Escherichia">de Wet, J. R., Wood, K. V., Helinski, D. R., & DeLuca, M. (1985). Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America, 82(23), 7870–7873. https://doi.org/10.1073/pnas.82.23.7870</ref><ref name="Throne">Thorne, N., Shen, M., Lea, W. A., Simeonov, A., Lovell, S., Auld, D. S., & Inglese, J. (2012). Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter. Chemistry & biology, 19(8), 1060–1072. https://doi.org/10.1016/j.chembiol.2012.07.015</ref>, amongst other things. | ||
Revision as of 19:34, 27 April 2021
Firefly Luciferase
apple juice! B~)
| |||||||||||
References
- ↑ Branchini, B. R., Magyar, R. A., Murtiashaw, M. H., Anderson, S. M., Helgerson, L. C., & Zimmer, M. (1999). Site-directed mutagenesis of firefly luciferase active site amino acids: a proposed model for bioluminescence color. Biochemistry 38(40), 13223–13230. https://doi.org/10.1021/bi991181o
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Sundlov, J. A., Fontaine, D. M., Southworth, T. L., Branchini, B. R., Gulick, A. M. (2012). Crystal Structure of Firefly Luciferase in a Second Catalytic Conformation Supports a Domain Alternation Mechanism. Biochemistry 51(33), 6493-6495. https://doi.org/10.1021/bi300934s
- ↑ Marahiel, M. A., Stachelhaus, T., Mootz, H. D. (1997). Modular Peptide Synthetases Involved in Nonribosmal Peptide Synthesis. Chemical Reviews 97(7), 2651-2674. https://doi.org/10.1021/cr960029e
- ↑ 4.0 4.1 Branchini, B. R., Southworth, T. L., Murtiahsaw, M. H., Wilkinson, S. R., Khattak, N. F., Rosenberg, J. C., & Zimmer, M. (2005). Mutagenesis Evidence that the Partial Reactions of Firefly Bioluminescence are Catalyzed by Different Conformations of the Luciferase C-Terminal Domain. “Biochemistry 44”(5), 1385-1393. https://doi.org/10.1021/bi047903f
- ↑ Oba, Y., Ojika, M., Inouye, S. (2003). Firefly luciferase is a bifunctional enzyme: ATP-dependent monoxygenase and a long chain fatty acyl-CoA synthetase. “FEBS Letters 540”(1-3), 251-254. https://doi.org/10.1016/S0014-5793(03)00272-2
- ↑ Nakamura, M., Maki, S., Amano, Y., Ohkita, Y., Niwa, K., Hirano, T., Ohmiya, Y., & Niwa, H. (2005). Firefly luciferase exhibits bimodal action depending on the luciferin chirality. “Biochemical and Biophysical Research Communications, 331”(2), 471–475. https://doi.org/10.1016/j.bbrc.2005.03.202
- ↑ Branchini, B. R., Murtiashaw, M. H., Magyar, R. A., Anderson, S. M. (2000). The Role of Lysine 529, a Conserved Residue of the Acyl-Adenylate-Forming Enzyme Superfamily, in Firefly Luciferase. Biochemistry 39(18), 5433-5440. https://doi.org/10.1021/bi9928804
- ↑ Sala-Newby, G. B., & Campbell, A. K. (1991). Engineering a bioluminescent indicator for cyclic AMP-dependent protein kinase. “The Biochemical Journal”, 279 (Pt 3), 727–732. https://doi.org/10.1042/bj2790727
- ↑ de Wet, J. R., Wood, K. V., DeLuca, M., Helinski, D. R., & Subramani, S. (1987). Firefly luciferase gene: structure and expression in mammalian cells. Molecular and cellular biology, 7(2), 725–737. https://doi.org/10.1128/mcb.7.2.725
- ↑ de Wet, J. R., Wood, K. V., Helinski, D. R., & DeLuca, M. (1985). Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America, 82(23), 7870–7873. https://doi.org/10.1073/pnas.82.23.7870
- ↑ Thorne, N., Shen, M., Lea, W. A., Simeonov, A., Lovell, S., Auld, D. S., & Inglese, J. (2012). Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter. Chemistry & biology, 19(8), 1060–1072. https://doi.org/10.1016/j.chembiol.2012.07.015