We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Leanne Price/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 22: | Line 22: | ||
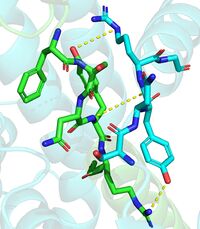
| - | The DGAT1 dimer structure is formed primarily through many [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen-bonding] interactions between the first 20 resolved residues (His69-Gly87). Hydrophobic interactions of the transmembrane helix region (Phe82-Ile98) with the other monomer also support the dimer structure formation. Additionally, there are four phospholipids present at the dimer interface that have been thought to contribute to the interactions between DGAT1 monomers. <ref name="Wang" /> | + | The DGAT1 dimer structure is formed primarily through many [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen-bonding] interactions between the first 20 resolved residues (His69-Gly87)(see figure 4). Hydrophobic interactions of the transmembrane helix region (Phe82-Ile98) with the other monomer also support the dimer structure formation. Additionally, there are four phospholipids present at the dimer interface that have been thought to contribute to the interactions between DGAT1 monomers. <ref name="Wang" /> |
| + | [[Image:Hbond.jpg|200 px|left|thumb|Figure 4. Shows the interactions between residues His69-Gly87. One dimmer is shown in blue and the other in green(6vyi). ]] | ||
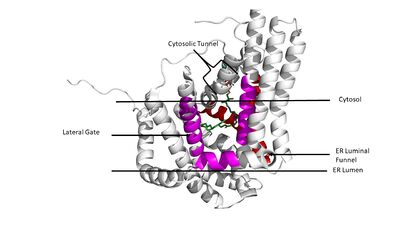
===Tunnels=== | ===Tunnels=== | ||
| Line 37: | Line 38: | ||
==Mechanism== | ==Mechanism== | ||
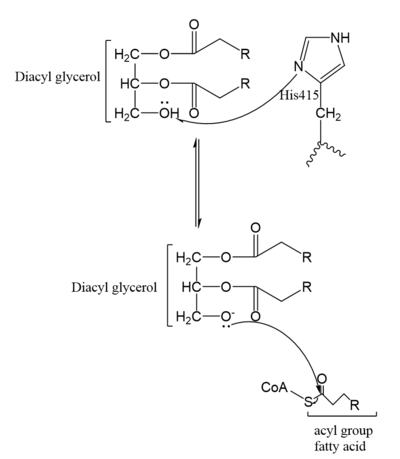
| - | [[Image:Mechanism.PNG|400 px|left|thumb|Figure | + | [[Image:Mechanism.PNG|400 px|left|thumb|Figure 5. Mechanism for DGAT1]] |
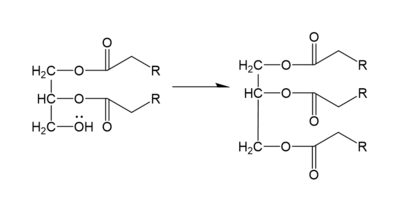
In the DGAT1 mechanism (Figure 4), the diglyceride serves as the nucleophile. While the acyl group of the CoA enzyme serves as the electrophile. The lone pair on the last hydroxyl group present on the glycerol of the diglyceride attacks the thioester bond of the acyl-CoA enzyme. This attack breaks the sulfur-carbon bond, a weak bond that is easily breakable. This allows the acyl group of the acyl-CoA enzyme to attach to the diglyceride, creating a triglyceride. While the CoA group then serves as the leaving group. | In the DGAT1 mechanism (Figure 4), the diglyceride serves as the nucleophile. While the acyl group of the CoA enzyme serves as the electrophile. The lone pair on the last hydroxyl group present on the glycerol of the diglyceride attacks the thioester bond of the acyl-CoA enzyme. This attack breaks the sulfur-carbon bond, a weak bond that is easily breakable. This allows the acyl group of the acyl-CoA enzyme to attach to the diglyceride, creating a triglyceride. While the CoA group then serves as the leaving group. | ||
Revision as of 20:47, 27 April 2021
Diacylglycerol O-Acyltransferase 1
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Sui X, Wang K, Gluchowski NL, Elliott SD, Liao M, Walther TC, Farese RV Jr. Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature. 2020 May;581(7808):323-328. doi: 10.1038/s41586-020-2289-6. Epub 2020 May, 13. PMID:32433611 doi:http://dx.doi.org/10.1038/s41586-020-2289-6
- ↑ 2.0 2.1 Wang L, Qian H, Nian Y, Han Y, Ren Z, Zhang H, Hu L, Prasad BVV, Laganowsky A, Yan N, Zhou M. Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature. 2020 May;581(7808):329-332. doi: 10.1038/s41586-020-2280-2. Epub 2020 May, 13. PMID:32433610 doi:http://dx.doi.org/10.1038/s41586-020-2280-2
- ↑ https://www.proteinatlas.org/ENSG00000185000-DGAT1/pathology
Student Contributors
- Justin Smith
- Eloi Bigirimana
- Leanne Price