User:Hannah Wright/Sandbox 1
From Proteopedia
< User:Hannah Wright(Difference between revisions)
| Line 34: | Line 34: | ||
The <scene name='87/877636/Calcium_ion_coordination/2'>calcium ion</scene> has been shown to convert inactive LPL to the active dimer form. The calcium ion is coordinated by residues A194, R197, S199, D201, and D202. Mutations in the coordinating residues can give rise to detrimental metabolic diseases<ref name="Birrane">PMID:30559189</ref>. The crystal structures of LPL revealed that the carboxylic acid side chain of D201 significantly aids in the coordination of LPL with the calcium ion. If D201 is mutated to a valine, for example, LPL can no longer fold correctly, and thus, LPL secretion from cells is inhibited due to the loss of the carboxylic acid side chain <ref name="Birrane">PMID:30559189</ref>. | The <scene name='87/877636/Calcium_ion_coordination/2'>calcium ion</scene> has been shown to convert inactive LPL to the active dimer form. The calcium ion is coordinated by residues A194, R197, S199, D201, and D202. Mutations in the coordinating residues can give rise to detrimental metabolic diseases<ref name="Birrane">PMID:30559189</ref>. The crystal structures of LPL revealed that the carboxylic acid side chain of D201 significantly aids in the coordination of LPL with the calcium ion. If D201 is mutated to a valine, for example, LPL can no longer fold correctly, and thus, LPL secretion from cells is inhibited due to the loss of the carboxylic acid side chain <ref name="Birrane">PMID:30559189</ref>. | ||
===Active Site=== | ===Active Site=== | ||
| - | The <scene name='87/878236/Active_site_4_20/4'>active site</scene> of LPL is composed of multiple pieces. The <scene name='87/878236/Active_site_4_20/5'>hydrophobic entry</scene> to the binding site outlines the general structure and provides considerable stability to the active site. The <scene name='87/878236/Active_site_lid_region_fixed/6'>lid region</scene> is also an important component of the active site, occupying residues 243-266, which is vital for the recognition of substrates. The <scene name='87/878236/Active_site_w_catalytic_residu/7'>catalytic residues</scene> consist of the [http://en.wikipedia.org/wiki/Catalytic_triad#:~:text=A%20catalytic%20triad%20is%20a,lipases%20and%20%CE%B2%2Dlactamases) catalytic triad] (residues H268, S159 and D183) and the [http://en.wikipedia.org/wiki/Oxyanion_hole#:~:text=An%20oxyanion%20hole%20is%20a,amides%20or%20positively%20charged%20residues oxyanion hole], consisting of residues L160 and W82. The catalytic residues catalyze the reaction of the typical substrate of LPL, triglycerides, and the oxyanion hole is responsible for aiding in the stability of the transition state of substrates. The main chain nitrogens stabilize the tetrahedral intermediate <ref name="Birrane">PMID:30559189</ref>. | + | The <scene name='87/878236/Active_site_4_20/4'>active site</scene> of LPL is composed of multiple pieces. The <scene name='87/878236/Active_site_4_20/5'>hydrophobic entry</scene> to the binding site outlines the general structure and provides considerable stability to the active site. The <scene name='87/878236/Active_site_lid_region_fixed/6'>lid region</scene> is also an important component of the active site, occupying residues 243-266, which is vital for the recognition of substrates. The <scene name='87/878236/Active_site_w_catalytic_residu/7'>catalytic residues</scene> consist of the [http://en.wikipedia.org/wiki/Catalytic_triad#:~:text=A%20catalytic%20triad%20is%20a,lipases%20and%20%CE%B2%2Dlactamases) catalytic triad] (residues H268, S159 and D183) and the [http://en.wikipedia.org/wiki/Oxyanion_hole#:~:text=An%20oxyanion%20hole%20is%20a,amides%20or%20positively%20charged%20residues oxyanion hole], consisting of residues <scene name='87/877636/Activesite_oxyanionhole/4'>L160 and W82</scene>. The catalytic residues catalyze the reaction of the typical substrate of LPL, triglycerides, and the oxyanion hole is responsible for aiding in the stability of the transition state of substrates. The main chain nitrogens stabilize the tetrahedral intermediate <ref name="Birrane">PMID:30559189</ref>. |
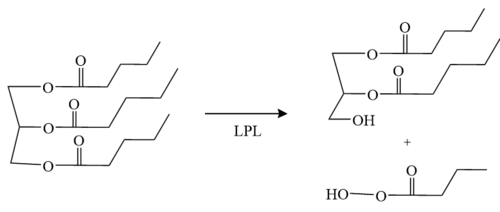
====Mechanism==== | ====Mechanism==== | ||
Current revision
Lipoprotein Lipase (LPL) complexed with GPIHBP1
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116
- ↑ Davies BS, Beigneux AP, Barnes RH 2nd, Tu Y, Gin P, Weinstein MM, Nobumori C, Nyren R, Goldberg I, Olivecrona G, Bensadoun A, Young SG, Fong LG. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 2010 Jul 7;12(1):42-52. doi: 10.1016/j.cmet.2010.04.016. PMID:20620994 doi:http://dx.doi.org/10.1016/j.cmet.2010.04.016
- ↑ 3.0 3.1 Wong H, Davis RC, Thuren T, Goers JW, Nikazy J, Waite M, Schotz MC. Lipoprotein lipase domain function. J Biol Chem. 1994 Apr 8;269(14):10319-23. PMID:8144612
- ↑ Arora R, Nimonkar AV, Baird D, Wang C, Chiu CH, Horton PA, Hanrahan S, Cubbon R, Weldon S, Tschantz WR, Mueller S, Brunner R, Lehr P, Meier P, Ottl J, Voznesensky A, Pandey P, Smith TM, Stojanovic A, Flyer A, Benson TE, Romanowski MJ, Trauger JW. Structure of lipoprotein lipase in complex with GPIHBP1. Proc Natl Acad Sci U S A. 2019 May 21;116(21):10360-10365. doi:, 10.1073/pnas.1820171116. Epub 2019 May 9. PMID:31072929 doi:http://dx.doi.org/10.1073/pnas.1820171116
- ↑ Beigneux AP, Davies BS, Gin P, Weinstein MM, Farber E, Qiao X, Peale F, Bunting S, Walzem RL, Wong JS, Blaner WS, Ding ZM, Melford K, Wongsiriroj N, Shu X, de Sauvage F, Ryan RO, Fong LG, Bensadoun A, Young SG. Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 2007 Apr;5(4):279-91. doi: 10.1016/j.cmet.2007.02.002. PMID:17403372 doi:http://dx.doi.org/10.1016/j.cmet.2007.02.002
- ↑ 6.0 6.1 Falko JM. Familial Chylomicronemia Syndrome: A Clinical Guide For Endocrinologists. Endocr Pract. 2018 Aug;24(8):756-763. doi: 10.4158/EP-2018-0157. PMID:30183397 doi:http://dx.doi.org/10.4158/EP-2018-0157
Student/Contributors
- Ashrey Burley
- Allison Welz
- Hannah Wright