User:Giselle Flores/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
==Structural Overview== | ==Structural Overview== | ||
===LPL=== | ===LPL=== | ||
| - | <scene name='87/877513/Original_scene/1'>LPL</scene> is assumed to only be active as a <scene name='87/877513/Lpl_dimer/ | + | <scene name='87/877513/Original_scene/1'>LPL</scene> is assumed to only be active as a <scene name='87/877513/Lpl_dimer/5'>tetramer</scene> composed of two LPL-GPIHBP1 heterodimers, however, previous studies have argued that the lipase can be active in its <scene name='87/877513/Original_scene/1'>single heterodimeric form</scene>.<ref name=”Arora”>PMID:31072929</ref><ref name=”Beigneux”>PMID:30850549</ref>The N-terminal domain of lipoprotein lipase is known to consist of an alpha/beta hydrolase domain, which is composed of six alpha helices and ten beta-strands. This domain creates an <scene name='87/877513/Alpha-beta_hydrolase_domain_1/3'>alpha beta hydrolase fold</scene>. The C-terminal domain of lipoprotein lipase is composed of twelve beta strands which form a "<scene name='87/877513/Just_barrel_domain_1/1'>barrel domain</scene>".<ref name=”Arora”>PMID:31072929</ref> |
=== GPIHBP1 === | === GPIHBP1 === | ||
| - | Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 ([https://en.wikipedia.org/wiki/GPIHBP1 GPIHBP1]) is a secondary domain that is critical to the stabilization, function, and movement of <scene name='87/877513/Original_scene/1'>LPL</scene>.<ref name=”Birrane”>PMID:30559189</ref> The GPIHBP1’s highly acidic and intrinsically disordered N-terminal domain are essential to the binding of LPL’s C-terminal Domain. It has been shown that GPIHBP1 has | + | Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 ([https://en.wikipedia.org/wiki/GPIHBP1 GPIHBP1]) is a secondary domain that is critical to the stabilization, function, and movement of <scene name='87/877513/Original_scene/1'>LPL</scene>.<ref name=”Birrane”>PMID:30559189</ref> The GPIHBP1’s highly acidic and intrinsically disordered N-terminal domain are essential to the binding of LPL’s C-terminal Domain. It has been shown that GPIHBP1 has a “three fingered domain”, which holds it tightly to LPL by <scene name='87/877513/Hydrophobic_interface-labeled/3'>hydrophobic interactions</scene>.<ref name=”Birrane”>PMID:30559189</ref> The importance of GPIBP1’s affinity to LPL was analyzed by Birrane et al.<ref name=”Birrane”>PMID:30559189</ref>, and it was found that missense mutations of critical residues resulted in high amounts of impairments. It was also concluded that these impairments caused hypertriglyceridemia (chylomicronemia).<ref name=”Birrane”>PMID:30559189</ref> |
== Structural Highlights == | == Structural Highlights == | ||
| Line 17: | Line 17: | ||
=== Lid and Lipid Binding Region === | === Lid and Lipid Binding Region === | ||
| - | In the presence of the GPIHBP1 inhibitor, the <scene name='87/877514/Lid_region_final/1'>Lid Region</scene> and lipid-binding region become visible within the structure. As displayed through a study conducted by Arora et. al, in 2019, the lipid-binding region of LPL actively interacts with the known inhibitor in the dimeric form. <ref name=”Arora”>PMID:31072929</ref> This was established to be the only time that the | + | In the presence of the GPIHBP1 inhibitor, the <scene name='87/877514/Lid_region_final/1'>Lid Region</scene> and <scene name='87/877516/Inhibitorsbound/1'>lipid-binding region</scene> become visible within the structure. As displayed through a study conducted by Arora et. al, in 2019, the lipid-binding region of LPL actively interacts with the known inhibitor in the dimeric form. <ref name=”Arora”>PMID:31072929</ref> This was established to be the only time that the heterodimeric form was shown as an active lipase. The lid region residues Ile245, Ile249, V251, Ile252, Leu257, Val260, Leu263, and Val264, are found as an open conformation which is composed of two small alpha helices that reach out and away from the protein. The lid and lipid-binding region create hydrophobic patches on the surface of lipoprotein lipase which are essential for <scene name='87/877514/Lipid_binding_and_lid/1'>ligand binding</scene> by LPL. |
| - | [[Image:Inhibiting.png|300 px|right|thumb|The novel inhibitor bound between the lipid-binding region of one LPL | + | [[Image:Inhibiting.png|300 px|right|thumb|The novel inhibitor bound between the lipid-binding region of one LPL and the catalytic site of the other LPL of the tetramer.]] |
==Mechanism== | ==Mechanism== | ||
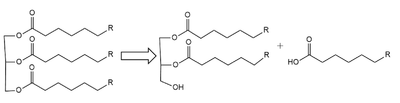
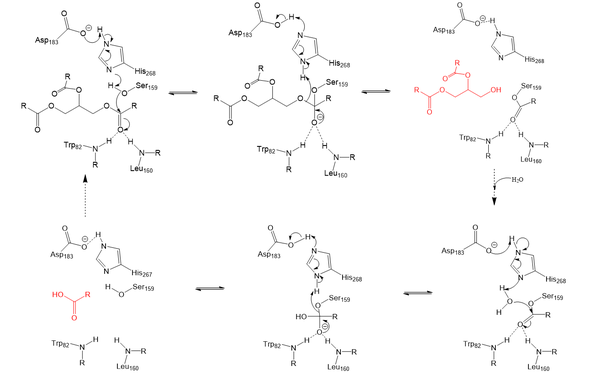
| - | Lipoprotein Lipase functions to catalyze the hydrolysis of one [https://en.wikipedia.org/wiki/Ester ester bond] of triglycerides in order to remove one fatty acid tail and turn the triglyceride into a diglyceride. It does this by utilizing a simple [https://en.wikipedia.org/wiki/Serine_hydrolase serine hydrolase] mechanism, in which it uses a <scene name='87/877513/Catalytic_triad-1/6'>catalytic triad</scene> composed of Asp183, His268, and Ser159 to catalyze the hydrolysis. His268 serves as a base catalyst by deprotonation of Ser159, which can then serve as the [https://en.wikipedia.org/wiki/Nucleophile nucleophile] | + | Lipoprotein Lipase functions to catalyze the hydrolysis of one [https://en.wikipedia.org/wiki/Ester ester bond] of triglycerides in order to remove one fatty acid tail and turn the triglyceride into a diglyceride. It does this by utilizing a simple [https://en.wikipedia.org/wiki/Serine_hydrolase serine hydrolase] mechanism, in which it uses a <scene name='87/877513/Catalytic_triad-1/6'>catalytic triad</scene> composed of Asp183, His268, and Ser159 to catalyze the hydrolysis. His268 serves as a base catalyst by deprotonation of Ser159, which can then serve as the [https://en.wikipedia.org/wiki/Nucleophile nucleophile] to attack the carbonyl carbon of one of the fatty acid chains of a triglyceride. This forms a tetrahedral intermediate, which is stabilized by the amide of Trp82 and Leu160 residues, called the <scene name='87/877513/Oxyanion_hole_-_labeled/8'>oxyanion hole</scene>. The single fatty acid chain is cleaved from the triglyceride, forming the diglyceride product. Then, water is used to free the fatty acid from Ser159 and return all of the residues back to their starting point, in order to perform the mechanism again on another triglyceride. Ultimately, the hydrolysis results in the formation of one free fatty acid and glycerol with two fatty acid tails (Figure 2). |
[[Image:LPL_final_Mechanism.png|600 px|center|thumb|Figure 2: Serine hydrolase mechanism utilized by LPL to catalyze the breakdown of one ester bond of a triglyceride. Compounds colored red are the products of the hydrolysis.]] | [[Image:LPL_final_Mechanism.png|600 px|center|thumb|Figure 2: Serine hydrolase mechanism utilized by LPL to catalyze the breakdown of one ester bond of a triglyceride. Compounds colored red are the products of the hydrolysis.]] | ||
| Line 27: | Line 27: | ||
===M404 Mutation=== | ===M404 Mutation=== | ||
| - | A 2018 study <ref name=”Paquette”>PMID:29452893</ref> argued that mutations that inhibit the proper binding of GPIHBP1 and LPL are suspected to cause chylomicronemia. A mutation that is believed to cause chylomicronemia is known as <scene name='87/877514/M404_mutation_site/1'>M404R</scene>. Within this mutation, the methionine is replaced with a large and hydrophilic arginine. While this mutation does not impact LPL secretion, it does affect the formation of the LPL-GPIHBP1 complex | + | A 2018 study <ref name=”Paquette”>PMID:29452893</ref> argued that mutations that inhibit the proper binding of GPIHBP1 and LPL are suspected to cause chylomicronemia. A mutation that is believed to cause chylomicronemia is known as <scene name='87/877514/M404_mutation_site/1'>M404R</scene>. Within this mutation, the methionine is replaced with a large and hydrophilic arginine. While this missense mutation does not impact LPL secretion, it does affect the formation of the LPL-GPIHBP1 complex by disrupting LPL’s interaction with GPIHB1’s V121, E122, T124, and V126 residues. This disrupts the stabilization of the LPL-GPIHBP1 complex, which has been seen to negatively effect LPL’s ability to catalyze the hydrolysis of triglycerides. |
| Line 40: | Line 40: | ||
<ref name=”Beigneux”>PMID:30850549</ref> | <ref name=”Beigneux”>PMID:30850549</ref> | ||
<ref name=”Mead”>PMID:12483461</ref> | <ref name=”Mead”>PMID:12483461</ref> | ||
| - | <ref name=”Eckel”>PMID:2648155</ref> | + | <ref name=”Eckel”>PMID:2648155</ref> |
<ref name=”Francis”>PMID:11905095</ref> | <ref name=”Francis”>PMID:11905095</ref> | ||
<ref name=”Austin”>PMID:9526807</ref> | <ref name=”Austin”>PMID:9526807</ref> | ||
| Line 58: | Line 58: | ||
Maggie Stopa | Maggie Stopa | ||
| - | |||
| - | |||
| - | <scene name='87/877514/Small_alpha_hel/1'>small alpha helix</scene> | ||
Revision as of 01:29, 28 April 2021
Lipoprotein Lipase coupled with GPIHBP1
| |||||||||||